Abstract
The aim of this study is to analyse the expression status of long non-coding RNA (lncRNA) H19 in nasopharyngeal carcinoma and to unravel its oncogenic properties at molecular level. The abundance of H19, let-7a, b, g, i and HRAS was quantified by real-time PCR. Cell viability was measured by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide method. Cell proliferation was evaluated by the cell counting. Cell migration and cell invasion were determined using transwell chamber and scattering colony formation. Tumour progression was monitored in xenograft tumour model and tail vein injection was adopted for lung metastasis assessment. Luciferase reporter assay was employed to interrogate the potential regulatory action of let-7 genes on H19 expression. The endogenous HRAS protein was quantified by western blotting. H19 was aberrantly over-expression in nasopharyngeal carcinoma, which intimately associated with poorer prognosis. H19-deficency significantly inhibited cell viability and suppressed cell proliferation. Furthermore, both migrative and invasive capacity were compromised by H19 knockdown. H19-silencing remarkably delayed xenograft tumour progression and lung metastasis. Mechanistically, H19 competitively sponged let-7 genes and therefore up-regulated HRAS, which consequently contributed to its oncogenic activity in nasopharyngeal carcinomas. Our study uncovered the oncogenic properties of H19 in nasopharyngeal carcinoma and highlighted the H19-let-7-HRAS signalling axis underlying the incidence and metastasis of this disease.
Introduction
Nasopharyngeal carcinoma is the most common cancer originating in the nasopharynx [Citation1]. The morbidity of nasopharyngeal carcinoma is relatively rare in most of the world, while extremely common in southern regions of China [Citation2]. Especially in Guangdong Province, nasopharyngeal carcinoma accounts for 18% of all cancer in China. In 2010, it is estimated that 65,000 deaths were claimed by this disease globally. The occurrence of nasopharyngeal carcinoma frequently relates to the combination of risk factors including virus infection, dietary habits and heredities. The infection with Epstein-Barr virus (EBV) is the unequivocal causal factor for this disease [Citation3]. The traditional dietary habit in southern China including mass consumption of salted vegetable, fish and meat and hot tea is considered critically contributing to morbidity of this Cantonese cancer [Citation4]. In addition, various mutations that over-activate NF-κB signalling have been identified in almost half of nasopharyngeal carcinomas. Clinically, nasopharyngeal carcinoma can be managed by surgery, chemotherapy and radiotherapy [Citation5], which heavily depends on the stage of tumour and overall health status. Recently, the unique expression of EBV latent protein in undifferentiated nasopharyngeal carcinoma emerged as a potent target for immunotherapeutic exploitations.
Long non-coding RNA (lncRNA) is a class of abundant transcripts longer than 200 nucleotides without protein coding potential. The multifaceted biological functions of lncRNA have been uncovered in regulation of gene transcription, post-transcriptional regulation and epigenetic regulations [Citation6]. Cumulative evidence implicated the critical roles in variety of human malignancies [Citation7]. In nasopharyngeal carcinoma, Bo et al. reported up-regulated lncRNA AFAP1-AS1 expression is associated with progression and poor prognosis [Citation8]. Sun et al. demonstrated that LET repressed by EZH2 inhibits cell proliferation and induces apoptosis of nasopharyngeal carcinoma cell [Citation9]. LOC401317, a p53-regulated lncRNA, has been reported to inhibit cell proliferation and induce apoptosis in the nasopharyngeal carcinoma cell line HNE2 [Citation10]. Along this direction, here we sought to characterize the expression status of H19 and unravel its mechanistic involvement in nasopharyngeal carcinoma.
LncRNA H19 features in the exclusive expression from the maternal allele. The product of this gene is increasingly recognized as oncogene in range of human cancers. For example, the study performed by Luan et al. reported that H19 promoted glucose metabolism and cell growth in malignant melanoma via miR-106a-5p/E2F3 axis [Citation11]. Liang et al. demonstrated that sTLR4/MD-2 complex inhibited colorectal cancer migration and invasiveness in vitro and in vivo by lncRNA H19 down-regulation [Citation12]. Yan et al. suggested that H19/miR-675 axis promoted gastric cancer via FADD/Caspase 8/Caspase 3 signalling pathway [Citation13]. In respect to nasopharyngeal carcinoma, Ng et al. disclosed the regulation of H19 imprinting gene expression by promoter hypomethylation [Citation14]. More importantly, Li et al. demonstrated that H19 regulated EZH2 expression by interacting with miR-630 and promoted cell invasion [Citation15]. However, the credibility of this study is evidently compromised by the sole experiment with cell culture in vitro. Therefore, here we sought to revisit this subject via employment of nasopharyngeal carcinoma xenograft mice model and prepared to unravel the underlying mechanisms.
Materials and methods
Cell culture
The human nasopharyngeal cancer cell lines CNE1 and CNE2 were purchased from the CellBank of Chinese Academy of Sciences (Beijing, China). All cells were regularly monitored for potential mycoplasma contamination. Cells were maintained in RPMI modified medium supplemented with 12% foetal bovine serum (FBS, Hyclone, Logan, UT) and 1% antibiotics mixture (Gibco, Grand Island, NY). Cells were cultured in humidified CO2 incubator (5%) and passaged at 1:4. For transfection, Lipofectamine 2000 (Invitrogen, Waltham, MA) was used in strict accordance with the manufacturer’s instruction.
Tumour samples
The nasopharyngeal tumour tissues and adjacent benign tissues were collected from the First Affiliated Hospital of Zhengzhou University from June 2016 to September 2017. The written informed consents were obtained from all enrolled patients. The study was approved by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University.
Real-time PCR
Quantitative PCR was performed with 7900HT Fast Real-Time PCR System (Applied Biosystems, Waltham, MA). RNA from indicated samples was isolated with RNeasy Mini Kit (Qiagen, Valencia, CA) following the provider’s manual. BioAnalyzer 2100 (Agilent, Santa Clara, CA) was employed to determine the quality and quantity of RNA samples prior to any further processing. The iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Hercules, CA) was used for cDNA preparation. The real-time PCR was performed with SYBR Green PCR Kit (Qiagen, Valencia, CA) and the primers were listed as below:
H19 forward primer: 5′-CACTGGCCTCCAGAGCCCGT-3′
H19 reverse primer: 5′-CGTCTTGGCCTTCGGCAGCTG-3′
GAPDH forward primer: 5′-GGAGCGAGATCCCTCCAAAAT-3′
GAPDH reverse primer: 5′-GGCTGTTGTCATACTTCTCATGG-3′.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay
The Vybrant MTT Cell Proliferation Assay Kit from ThermoFisher (Waltham, MA) was used to measure the cell viability in accordance with the manufacturer’s recommendation. Briefly, the H19-proficient and -deficient CNE1 and CNE2 cells were seeded into 96-well in triplicate and 24-h consecutive culture was allowed. The cells were replaced with 100 μL fresh medium and followed by the addition of 10 μL of MTT stock solution. The formazan was formed at 37 °C for 4 h and dissolved by 100 μL of SDS-HCl for another 4 h at 37 °C. The absorption at 570 nm was measured by Infinite F500 Microplate Reader (Tecan, Männedorf, Switzerland).
Transwell assay
The Corning Costar Transwell chamber (Corning, Corning, NY) was used to evaluate cell migration and invasion. For invasive assay, the carbonate membrane was pre-coated with 0.1% Matrigel (BD Biosciences, Franklin Lakes, NJ). The single-cell suspension in serum-free medium was added into the upper chamber, in which lower compartment was supplemented with 750 μL of complete medium with FBS as attractant. After 12 h, the unattached cells were cautiously removed with cotton swab. The invaded cells were fixed with paraformaldehyde (4%) first and then subjected to crystal violet (0.25%) staining for 15 min. The representative images were captured under light microscope and colony number was counted from three independent areas.
Scattering colony formation
The wild-type or H19-deficient cells were seeded into six-well plate in triplicate and subjected to continuous culture for one week. The colony formation was visualized by crystal violet staining. The morphology of colonies was categorized into scattered, loose and compact and determined under light microscope.
Xenograft mice
The immunodeficient NOD/SCID/IL2Rγ–/– (NSG) mice (3–4 weeks, 15–20 g) were ordered from VitalRiver (Beijing, China). All mice were housed in specific-pathogen-free (SPF) environment with free access to drinking water and food after acclimation for one week. The subcutaneous inoculation of cancer cells was adopted to establish xenograft tumour model. To investigate the metastasis potential, the indicated cells were i.v. injected into tail vein. All animal experiments were performed in strict accordance with the National Institutes of Health (NIH) guide and the protocol was approved by the Committee of Animal Use and Care of the First Affiliated Hospital of Zhengzhou University.
Ki-67 staining
The proliferative index was interrogated with Ki-67 staining. The mice were sacrificed at the endpoint of experiment and xenograft tumour was resected for paraffin-embedded tissue section. After brief antigen retrieval with 0.1 M sodium citrate (pH 6.0), the sections were incubated with mouse anti-Ki67 antibody (ab15580, Abcam, Cambridge, MA) at 4 °C overnight. The endogenous peroxidase was quenched by 0.3% H2O2 for 15 min and followed by HRP-conjugated secondary antibody incubation for 1 h at room temperature. The sections were developed in DAB solution and the representative images were acquired under light microscope.
Luciferase reporter assay
To investigate the potential regulatory effects of let-7 family on H19 expression, here, we subclone H19 into pSi-Check2 luciferase reporter plasmid (Promega, Madison, WI). CNE1 cells were co-transfected with Psi-Check2-H19 and let-t family members for 24 h; the relative luciferase activity was determined in the cell lysate with Bright-Glo Luciferase Assay System (Promega, Madison, WI) in accordance with manufacturer’s instruction. The measurement of luciferase intensity was performed on Infinite F500 Microplate Reader (Tecan, Männedorf, Switzerland).
Statistical analysis
Data were obtained from at least three independent repeats unless stated. Data processing and analysis was performed with GraphPad 7.0 (GraphPad Software, La Jolla, CA). SPSS 23.0 (SPSS Inc., Chicago, IL) (Student’s t test, Chi-square test, one- or two-way ANOVA analysis followed by a post hoc test) was employed for statistical comparison. The significance of p values was calculated and p < .05 was considered as significantly different.
Results
lncRNA H19 is unregulated in nasopharyngeal carcinoma and indicates a poor survival outcome
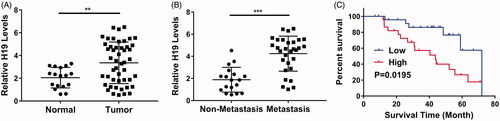
We first analysed the relative expression of H19 in the clinical nasopharyngeal carcinoma tissue samples. As shown in , the significant up-regulation of H19 in tumour was observed in comparison with adjacent normal counterparts. Further examination clearly demonstrated the higher abundance of H19 transcripts in cancer patient with lymph node metastasis compared with the non-metastatic ones (). Consistent with over-expression of H19 in nasopharyngeal carcinoma, Kaplan–Meier’s survival analysis exhibited the remarkable prolongation of the overall survival periods in the patients with low H19, while high level of H19 significantly associated with poorer clinical outcome (). Our data uncovered the aberrant over-expression of H19 in nasopharyngeal carcinoma, which predicted unfavourable prognosis.
Figure 1. lncRNA H19 is unregulated in nasopharyngeal carcinoma and indicates a poor survival outcome. (A) The H19 levels in normal adjacent tissues or tumours were determined by qPCR. (B) The H19 levels in patients with or without metastasis were determined by qPCR. (C) Overall survival analysis revealed that nasopharyngeal carcinoma patients with high H19 levels displayed poor survival outcomes. **p < .01, ***p < .001. Student’s t-test in (A) and (B), log-rank test in (C).
lncRNA H19 promotes nasopharyngeal carcinoma cells proliferation
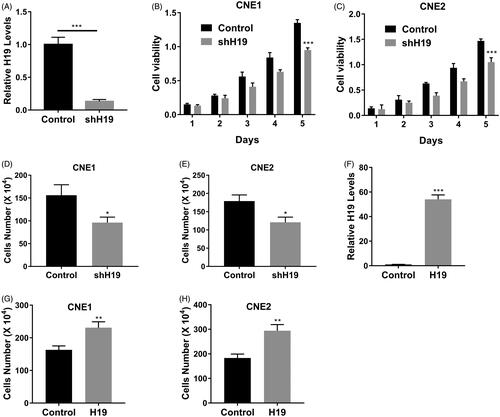
Next, we sought to investigate the potential impact of H19 expression on the proliferation of nasopharyngeal carcinoma cells. To this end, we specifically silenced H19 transcripts with shRNA, and the success in efficient knockdown of H19 is shown in . Cell viability interrogated with MTT assay was tremendously compromised in H19-deficient CNE1 () and CNE2 (). Likewise, the cell counting results demonstrated that H19-silencing greatly inhibited cell proliferation in CNE1 () and CNE2 (). On the contrary, ectopic over-expression of H19 () stimulated cell growth in both cell lines (). Our results uncovered the oncogenic properties of H19 in promoting nasopharyngeal carcinoma cell proliferation.
Figure 2. LncRNA H19 promotes nasopharyngeal carcinoma cells proliferation. (A) The mRNA expression of H19 in CNE1 cells transfected with H19 shRNA was determined by qPCR. (B) Cell viability of CNE1 cells transfected with H19 shRNA or vector was determined by MTT assay. (C) Cell viability of CNE2 cells transfected with H19 shRNA or vector was determined by MTT assay. (D) Cell viability of CNE1 cells transfected with H19 shRNA or vector was determined by cell counts assay. (E) Cell viability of CNE2 cells transfected with H19 shRNA or vector was determined by cell counts assay. (F) The mRNA expression of H19 in CNE1 cells transfected with H19 expression plasmid was determined by qPCR. (G) Cell viability of CNE1 cells transfected with H19 expression plasmid or vector was determined by cell counts assay. (H) Cell viability of CNE2 cells transfected with H19 expression plasmid or vector was determined by cell counts assay. Data are mean + SD. *p < .05, **p < .01 and ***p < .001.
LncRNA H19 promotes migration and invasion in nasopharyngeal carcinoma cells
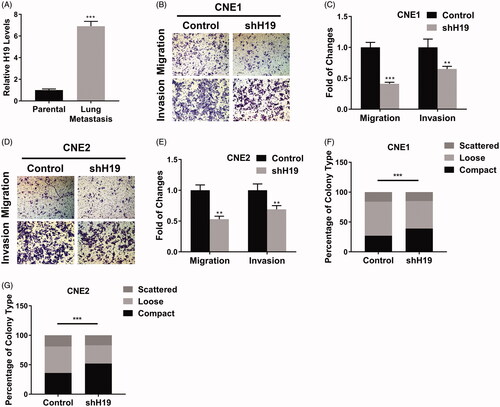
Our previous data indicated that high level of H19 related to lymph node metastasis as well. Here, we first experimentally confirmed this observation in the metastatic tumour mice via tail vein injection of CNE1 cells. The comparison of H19 contents in parental cells and lung-metastasized H19 offspring was performed with real-time PCR. As shown in , the tremendous up-regulation of H19 in lung metastasis was observed, which implicated a critical role of H19 for cancer spread. The invasive and migrative capacity of nasopharyngeal carcinoma in response to H19 manipulations was interrogated by transwell assay. Both invasion and migration were compromised in H19-deficient CNE1 cells (). Similarly, H19-knockdown in CNE2 cells significantly decreased the migrative and invasive potentials (). Furthermore, we performed the scattering colony formation assay to evaluate the impact of H19-silencing on the metastatic capacity of nasopharyngeal carcinoma cells. The percentage of compact colony was remarkably higher in H19-deficient cells in comparison with parental cells (). Therefore, our data indicated that high expression of H19 evidently contributed to the metastatic properties of nasopharyngeal carcinoma both in vivo and in vitro.
Figure 3. LncRNA H19 promotes migration and invasion in nasopharyngeal carcinoma cells. (A) The H19 levels in parental or lung-metastatic CNE1 cells were determined by qPCR. (B) Transwell migration and invasion assay of CNE1 cells transfected with H19 shRNA. (C) The statistical results of transwell migration and invasion assay in CNE1 cells. (D) Transwell migration and invasion assay of CNE2 cells transfected with H19 shRNA. (E) The statistical results of transwell migration and invasion assay in CNE2 cells. (F) Scattering colony formation assay of CNE1 cells transfected with shH19 or vector. (G) Scattering colony formation assay of CNE2 cells transfected with shH19 or vector. Data are mean + SD. **p < .01 and ***p < .001.
LncRNA H19 promotes nasopharyngeal carcinoma proliferation and metastasis in vivo
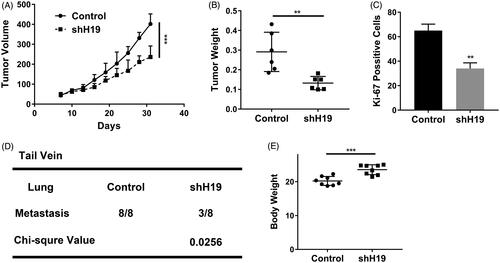
Our previous in vitro results suggested the intimate association between H19 and cell proliferation/metastasis in nasopharyngeal carcinoma. To exclude the potential artefacts originated from cell culture, we sought to validate this phenomenon in vivo with xenograft model. As shown in , H19-knockdown remarkably delayed xenograft tumour progression in nude mice during our experiment window. Consistently, the wet weights of xenograft tumour resected from the sacrifices mice at the endpoint were much higher in H19-silencing group than control (). The proliferative index indicated by the Ki67 staining manifested significantly inhibitory effects elicited by H19-knockdown (). Meanwhile, the lung metastasis potential of nasopharyngeal carcinoma was greatly inhibited in the H19-deficient cells, and three out of eight tail vein injected mice were characterized with metastatic sites in comparison with 8/8 in H19-proficient group (). Furthermore, we noticed the slight body-weight gain in H19-silencing mice, which definitely indicated more healthy status might be attributed to H19 deficiency (). In summary, here, we demonstrated the critical role of H19 in either tumour progression or metastasis in vivo.
Figure 4. LncRNA H19 promotes nasopharyngeal carcinoma proliferation and metastasis in vivo. (A) Tumour growth curve of CNE1 cells transfected with shH19 or vector. (B) The weight of tumours derived from CNE1 cells transfected with shH19 or vector. (C) Ki-67 staining of the tumours derived from CNE1 cells transfected with shH19 or vector. (D) The statistical of the lung metastasis in the tail veil injection mouse with CNE1 cells transfected with shH19 or vector. (E) The body weight of the tail vein injection mouse. Data are mean + SD. **p < .01 and ***p < .001.
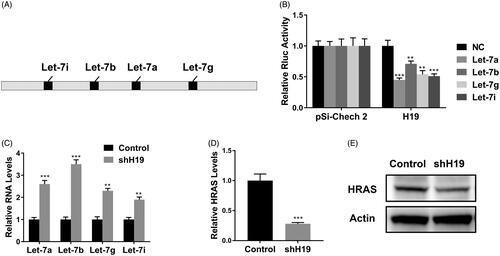
LncRNA H19 binds to let-7 family and regulates HRAS expression
Next, we sought to elucidate the molecular mechanism underlying the pro-proliferative and pro-metastatic properties of H19 in nasopharyngeal carcinoma. Close inspection of H19 sequence with RNAhybrid software has identified four let-7 family recognizing consensus motifs as illustrated in . To address the potential binding and regulatory action of let-7 family on H19, here we constructed H19-driven luciferase reporter vector. Co-transfection with let-7 family members including let-7a, let-7b, let-7g and let-7i markedly inhibited the luciferase activity in CNE1 cells (), which provided the direct evidence that let-7 family members recognized and modulated H19 expression. On the contrary, we observed the endogenous let-7a, let-7b, let-7g and let-7i were tremendously up-regulated in the H19-deficient CNE1 cells (), which implicated the suppressive effects of H19 on let-7 expression. In view of the well-acknowledged negative regulation of HRAS by let-7 family, we further examined the endogenous level of HRAS in response to H19 knockdown. As shown in , both mRNA and protein of HRAS were remarkably suppressed in H19-silenced CNE1 cells, which was in support of the competing sponge role of H19 upon let-7 family genes.
Figure 5. LncRNA H19 binds to let-7 family and regulates HRAS expression. (A) Schematic diagram of let-7 family binding sites on H19. (B) Luciferase reporter assay of pSi-check2 plasmid contains H19 and transfected with let-7 family. (C) The mRNA levels of let-7 family were determined in CNE1 cells transfected with shH19. (D) The HRAS levels in CNE1 cells transfected with shH19 were determined by qPCR. (E) The HRAS levels in CNE1 cells transfected with shH19 were determined by western blot. Data are mean + SD. **p < .01 and ***p < .001.
Discussion
The aberrant over-expression of H19 was well-documented in gastric [Citation16], colorectal [Citation17] and hepatocellular carcinomas [Citation18] and fundamentally linked to the incidence and development of these human malignancies. However, the relative expression status and mechanistic involvement of H19 in nasopharyngeal carcinoma were still elusive currently. Here, we characterized the up-regulation of H19 in nasopharyngeal carcinoma in comparison with adjacent benign tissues. We also demonstrated that aberrant high level of H19 is intimately associated with lung metastasis of nasopharyngeal carcinoma. Furthermore, the abundant H19 transcripts significantly predicted the unfavourable prognosis in our nasopharyngeal cancer panel. ShRNA-mediated knockdown of H19 in both CNE1 and CNE2 cells remarkably inhibited the cell viability and suppressed cell proliferation. On the contrary, ectopic introduction of exogenous H19 promoted cell growth in both cell lines. In addition, we observed tremendous increase of H19 transcripts in lung metastasis in comparison with parental CNE1 cells, which implicated the pro-metastatic property of H19 in nasopharyngeal cancer. Consistent with this notion, H19-deficiency greatly compromised cell migrative and invasive capacity, and resulted into more compact colonies in scattering colony formation assay. Our in vivo xenograft mice model consolidated this phenomenon, wherein deficiency in H19 significantly delayed tumour progression and associated with suppressed proliferative index. Simultaneously, H19-deficiency inhibited lung metastasis of tail vein-injected nasopharyngeal cancer cells. Mechanistically, we identified the putative consensus sequence of let-7 family genes on H19 transcript and experimentally validated the negatively regulatory effect of let-7 genes on H19 expression. In contrast, the endogenous contents of let-7 family genes were greatly increased in response to H19 knockdown. Most importantly, H19-silencing enhanced the inhibitory action of let-7 genes on HRAS, which consequently led to low level of HRAS and eventually the anti-tumour effect. Our data highlighted the critical role of H19-let-7-HRAS signalling in both incidence and metastasis of nasopharyngeal carcinoma, which might offer the opportunity for diagnostic, prognostic and therapeutic exploitations.
The competing endogenous RNA (ceRNA) has been proposed as one of the major mode of action for lncRNAs [Citation19]. Range of investigations has addressed the key roles of H19 so far in human malignancies functioning as ceRNA. For instance, Li et al. reported that lncRNA H19 promoted the proliferation and invasion of breast cancer through up-regulating DNMT1 expression by sponging miR-152 [Citation20]. Zhou et al. demonstrated that H19 mediated breast cancer cell plasticity during EMT and MET plasticity by differentially sponging miR-200b/c and let-7b [Citation21]. Yang et al. provided evidence that H19 promoted the migration and invasion of colon cancer by sponging miR-138 to up-regulate the expression of HMGA1 [Citation22]. In intestinal barrier, Su et al. showed H19 functioned as a ceRNA to regulate AQP3 expression by sponging miR-874 [Citation23]. The study performed by Liang et al. proposed H19 promoted epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer [Citation24]. Lv et al. reported that H19 regulated epithelial–mesenchymal transition and metastasis of bladder cancer [Citation25]. Zhao et al. displayed that H19 regulated ID2 expression through competitive binding to has-miR-19a/b in acute myelocytic leukaemia [Citation26]. Yang et al. uncovered that H19 promoted cell proliferation by competitively binding to miR-200a and de-repressing β-catenin expression in colorectal cancer [Citation17]. In thyroid cancer, Liu et al. demonstrated H19 competitively bound miR-17-5p to regulate YES1 expression [Citation27]. In addition to consensus sequence of let-7b, here, we identified target sites of let-7a, let-7g and let-7i on H19 transcript in nasopharyngeal carcinoma. More importantly, we unravelled that H19 indirectly up-regulated HRAS via competitive sponging let-7 genes, which consequently contributed to the oncology of nasopharyngeal cancer.
Noteworthily, despite the significant over-expression of H19 in nasopharyngeal carcinomas, the definite regulatory mechanism was still elusive currently and would be our priority in the future investigation. So far, several intriguing mechanisms have been proposed to underlie the up-regulation of H19 in variety of diseases. The investigation conducted by Ge et al. showed that VIGILIN involved in regulation of imprinting gene IGF2 and H19 in human hepatocellular carcinoma cell [Citation28]. Gao et al. disclosed the association of H19 promoter methylation with expression of H19 and IGF-II genes in adrenocortical tumours [Citation29]. Zhou et al. demonstrated that hypomethylation-mediated H19 overexpression increased the risk of disease evolution through the association with BCR-ABL transcript in chronic myeloid leukaemia [Citation30]. Wang et al. proposed that lncRNA H19 and HULC activated by oxidative stress promoted cell migration and invasion in cholangiocarcinoma through a ceRNA manner [Citation31]. Based on the established knowledge into dysregulation of H19, we speculated here that epigenetic modulation might play the predominant role in over-expression of H19 in nasopharyngeal carcinoma, which necessitated further clarifications.
Conclusions
In summary, we analysed the up-regulation of H19 in nasopharyngeal carcinomas and uncovered its oncogenic properties in respect to pro-proliferation and pro-metastasis effects. Our data highlighted the critical role of H19-let-7-HRAS signalling axis mechanistically in incidence and metastasis in this disease, which might be exploited for diagnostic and therapeutic purpose.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Chua MLK, Wee JTS, Hui EP, et al. Nasopharyngeal carcinoma. Lancet. 2016;387(10022):1012–1024.
- Wei KR, Zheng RS, Zhang SW, et al. Nasopharyngeal carcinoma incidence and mortality in China in 2010. Chin J Cancer. 2014;33:381–387.
- Dawson CW, Port RJ, Young LS. The role of the EBV-encoded latent membrane proteins LMP1 and LMP2 in the pathogenesis of nasopharyngeal carcinoma (NPC). Semin Cancer Biol. 2012;22(2):144–153.
- Liu Z, Chang ET, Liu Q, et al. Oral hygiene and risk of nasopharyngeal carcinoma—a population-based case-control study in China. Cancer Epidemiol Biomarkers Prev. 2016;25(8):1201–1207.
- Blanchard P, Lee A, Marguet S, et al. Chemotherapy and radiotherapy in nasopharyngeal carcinoma: an update of the MAC-NPC meta-analysis. Lancet Oncol. 2015;16(6):645–655.
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159.
- Maruyama R, Suzuki H. Long noncoding RNA involvement in cancer. BMB Rep. 2012;45(11):604–611.
- Bo H, Gong Z, Zhang W, et al. Upregulated long non-coding RNA AFAP1-AS1 expression is associated with progression and poor prognosis of nasopharyngeal carcinoma. Oncotarget. 2015;6(24):20404–20418.
- Sun Q, Liu H, Li L, et al. Long noncoding RNA-LET, which is repressed by EZH2, inhibits cell proliferation and induces apoptosis of nasopharyngeal carcinoma cell. Med Oncol. 2015;32(9):226.
- Gong Z, Zhang S, Zeng Z, et al. LOC401317, a p53-regulated long non-coding RNA, inhibits cell proliferation and induces apoptosis in the nasopharyngeal carcinoma cell line HNE2. PLoS One. 2014;9(11):e110674.
- Luan W, Zhou Z, Ni X, et al. Long non-coding RNA H19 promotes glucose metabolism and cell growth in malignant melanoma via miR-106a-5p/E2F3 axis. J Cancer Res Clin Oncol. 2018;144(3):531–542.
- Liang W, Zou Y, Qin F, et al. sTLR4/MD-2 complex inhibits colorectal cancer migration and invasiveness in vitro and in vivo by lncRNA H19 down-regulation. Acta Biochim Biophys Sin (Shanghai). 2017;49(11):1035–1041.
- Yan J, Zhang Y, She Q, et al. Long noncoding RNA H19/miR-675 axis promotes gastric cancer via FADD/Caspase 8/Caspase 3 signaling pathway. Cell Physiol Biochem. 2017;42(6):2364–2376.
- Ng A, Tang JP, Goh CH, et al. Regulation of the H19 imprinting gene expression in human nasopharyngeal carcinoma by methylation. Int J Cancer. 2003;104(2):179–187.
- Li X, Lin Y, Yang X, et al. Long noncoding RNA H19 regulates EZH2 expression by interacting with miR-630 and promotes cell invasion in nasopharyngeal carcinoma. Biochem Biophys Res Commun. 2016;473(4):913–919.
- Li H, Yu B, Li J, et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5(8):2318–2329.
- Yang W, Ning N, Jin X. The lncRNA H19 promotes cell proliferation by competitively binding to miR-200a and derepressing β-catenin expression in colorectal cancer. Biomed Res Int. 2017;2017:2767484.
- Wu J, Qin Y, Li B, et al. Hypomethylated and hypermethylated profiles of H19DMR are associated with the aberrant imprinting of IGF2 and H19 in human hepatocellular carcinoma. Genomics. 2008;91(5):443–450.
- Qi X, Zhang DH, Wu N, et al. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52(10):710–718.
- Li Z, Li Y, Li Y, et al. Long non-coding RNA H19 promotes the proliferation and invasion of breast cancer through upregulating DNMT1 expression by sponging miR-152. J Biochem Mol Toxicol. 2017;31(9):e21933.
- Zhou W, Ye XL, Xu J, et al. The lncRNA H19 mediates breast cancer cell plasticity during EMT and MET plasticity by differentially sponging miR-200b/c and let-7b. Sci Signal. 2017;10:eaak9557.
- Yang Q, Wang X, Tang C, et al. H19 promotes the migration and invasion of colon cancer by sponging miR-138 to upregulate the expression of HMGA1. Int J Oncol. 2017;50(5):1801–1809.
- Su Z, Zhi X, Zhang Q, et al. LncRNA H19 functions as a competing endogenous RNA to regulate AQP3 expression by sponging miR-874 in the intestinal barrier. FEBS Lett. 2016;590(9):1354–1364.
- Liang WC, Fu WM, Wong CW, et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6(26):22513–22525.
- Lv M, Zhong Z, Huang M, et al. lncRNA H19 regulates epithelial-mesenchymal transition and metastasis of bladder cancer by miR-29b-3p as competing endogenous RNA. Biochim Biophys Acta Mol Cell Res. 2017;1864(10):1887–1899.
- Zhao TF, Jia HZ, Zhang ZZ, et al. LncRNA H19 regulates ID2 expression through competitive binding to hsa-miR-19a/b in acute myelocytic leukemia. Mol Med Rep. 2017;16(3):3687–3693.
- Liu L, Yang J, Zhu X, et al. Long noncoding RNA H19 competitively binds miR-17-5p to regulate YES1 expression in thyroid cancer. FEBS J. 2016;283(12):2326–2339.
- Ge YJ, Xie XY, Yang B, et al. VIGILIN involves in regulation of imprinting gene IGF2 and H19 in human hepatocellular carcinoma cell. Sichuan Da Xue Xue Bao Yi Xue Ban. 2009;40(5):770–774.
- Gao ZH, Suppola S, Liu J, et al. Association of H19 promoter methylation with the expression of H19 and IGF-II genes in adrenocortical tumors. J Clin Endocrinol Metab. 2002;87(3):1170–1176.
- Zhou JD, Lin J, Zhang TJ, et al. Hypomethylation-mediated H19 overexpression increases the risk of disease evolution through the association with BCR-ABL transcript in chronic myeloid leukemia. J Cell Physiol. 2018;233(3):2444–2450.
- Wang WT, Ye H, Wei PP, et al. LncRNAs H19 and HULC, activated by oxidative stress, promote cell migration and invasion in cholangiocarcinoma through a ceRNA manner. J Hematol Oncol. 2016;9(1):117.
