Abstract
During transport of wastewater in force mains, sulphide and possibly methane formation take place due to prokaryotic activity. Sulphide has several detrimental effects and addition of ferrous or ferric iron for abatement by precipitation is commonly applied. Precipitation stoichiometry and efficiency of this process have been investigated in detail. However, it is largely unknown how ferrous and ferric iron influence prokaryotic populations of sewer biofilms. The microbiomes of iron-treated force main biofilms were, together with an untreated control, examined by sequencing of the 16S rDNA V3+ V4 regions. Differences in distribution and abundance of several bacterial and archaeal genera were observed, indicating that treatment with ferrous and ferric iron for sulphide abatement differentially changed sewer force main microbiomes. Furthermore, differences at the functional level (KEGG orthologs, KOs) indicate that ferrous and ferric iron treatment possibly can decrease methane formation, whereas functions related to dissimilatory sulphate reduction seemed unaffected.
Public Interest Statement
Sewer systems are an integral part of modern civilisation and little notice is given if everything works properly. In these underground structures microbes live by attaching themselves to pipe walls in slimy layers. These organisms decompose matters that are flushed down the drain and transported along the system. Among other substances, this microbial decomposition gives rise to sulphide. Sulphide has a distinct smell of rotten eggs, is extremely toxic, and is furthermore involved in premature failure of sewer systems. Greenhouse gasses such as methane can also arise from the decomposition taking place in these slimy layers. It is known how to render sulphide harmless by adding liquid iron to the sewage. But how iron addition influences the microbial community in the slimy layers and whether it has potential beneficial side effects in changing the ability of the microbes to develop sulphide and methane are investigated in this study.
Competing Interests
The authors declare no competing interests.
1. Introduction
During transport of wastewater in force mains, the available organic matter is degraded by microbial processes taking place mainly in the sewer biofilms. In these processes, oxidised substances that serve as electron acceptors are required. The order of utilisation of electron acceptors relevant for sewer systems is oxygen (O2), nitrate, ferric iron (Fe(III)), sulphate (SO42-), and carbon dioxide (Hvitved-Jacobsen, Vollertsen, & Nielsen, Citation2013). In force mains, oxygen, nitrate, and ferric iron are limited or quickly depleted due to microbial processes. Anaerobic conditions, therefore, prevail, where fermentative processes, sulphate reduction, and methane formation take place (Hvitved-Jacobsen et al., Citation2013). While methane emitted during transport of wastewater has drawn attention due to its greenhouse gas potential (Guisasola, de Haas, Keller, & Yuan, Citation2008), sulphate reduction is generally recognised as the main problem due to various issues related to the formed sulphide. The concentration of sulphide produced during wastewater transport has detrimental effects in terms of odour nuisance, corrosion of sewer assets, and health risk to sewage workers, all incurring substantial cost for handling (Apgar & Witherspoon, Citation2007; Boon, Citation1995).
The wastewater industry routinely abates sulphides to control these issues. A common method is sulphide precipitation using iron salts, where insoluble iron sulphides (FeS) are formed (ASCE, Citation1989; Ganigue, Gutierrez, Rootsey, & Yuan, Citation2011). The iron is typically dosed at the start of the force main, so it is available for precipitation when sulphide develops (Ganigue et al., Citation2011). Both ferrous (Fe(II)) and ferric iron can be used for the purpose. It is generally assumed that ferrous iron precipitates directly with sulphide, whereas ferric iron must undergo a reduction to ferrous iron before precipitating as FeS. During reduction to ferrous iron, sulphide is oxidised to elemental sulphur (S°) (ASCE, Citation1989). Precipitation of sulphide using ferrous iron is purely a chemical reaction. Ferric iron, on the other hand, must be reduced to ferrous iron either chemically with sulphide or biochemically before it can precipitate sulphide (Hvitved-Jacobsen et al., Citation2013; Lovley, Citation1991). In the biochemical pathway, ferric iron is used by chemoautotrophic bacteria, where the reduction is coupled with an oxidation of sulphide to elemental sulphur (Hvitved-Jacobsen et al., Citation2013). However, dissimilatory ferric iron reduction by some sulphate reducing bacteria can also be performed directly through enzymatic processes without the involvement of sulphide (Lovley, Roden, Phillips, & Woodward, Citation1993). A phylogenetic diverse group of bacteria has been found capable of performing dissimilatory ferric reduction, with some organisms being able to conserve energy from this process (Dworkin, Falkow, Rosenberg, Schleifer, & Stackebrandt, Citation2006). Fermentable sugars and amino acids can be oxidised and the resulting available fermentation end products such as acetate can also act as electron donors for ferric reduction by chemoheterotrophic bacteria (Dworkin et al., Citation2006; Hvitved-Jacobsen et al., Citation2013; Lovley, Citation1993). When common electron donors, such as acetate and hydrogen are scarce, ferric reducers have been shown to outcompete sulphate reducers and methane producers in sediments (Lovley & Phillips, Citation1987). However, in anoxic paddy soil it has been demonstrated that preferential use of one electron acceptor could be overcome (i.e. the use of Fe(III) over sulphate), by addition of electron donors to an otherwise electron donor depleted environment (Achtnich, Bak, & Conrad, Citation1995). This shows that these processes can mutually exclude each other or coexists in the same environment.
A study of anaerobic biofilms in a lab-scale simulated force main showed that ferric dosing inhibited sulphate reduction (Zhang, Keller, & Yuan, Citation2009), however, this finding was not linked directly to influences on the microbial population of the biofilm. A spatial variation of selected microbial species and activity of an untreated biofilm was found using a Robbins device setup, connected in parallel to a wastewater force main (Mohanakrishnan et al., Citation2009). Jensen et al. (Citation2016) in their study performed direct sampling in different sections of a sewer system and showed the spatial and temporal variability of bacterial communities. However, direct studies of bacterial communities in biofilms along the length of sewer force mains and how these communities respond to different stimuli are still largely lacking.
The objective of the current study was to assess the variation of microbial population in wastewater force mains due to treatment with ferrous and ferric iron for sulphide abatement. Emphasis was given to genera related to sulphide and methane production as well as ferric iron reducing microorganisms. The study was carried out on three parallel pilot-scale force mains pumping real wastewater and receiving ferrous ion, ferric ion, or no treatment. Biofilm samples were collected at different locations along the length of the three mains. Samples were analysed by using 16S rDNA sequencing to determine the sewer biofilm microbiomes.
2. Materials and methods
2.1. Force mains and biofilm sampling
Biofilms for microbiome analysis were collected from a sewer force main setup installed in a research and monitoring station in Frejlev, Denmark. An elaborate description of the system can be found in Kiilerich, Kiilerich, Nielsen, and Vollertsen (Citation2018). Briefly, the system consisted of three identical 300 m long full-flowing PE80 force mains of Ø50 mm, conveying fresh municipal wastewater. The mains were operated in parallel in an operational pattern mimicking real force mains. Biofilms were matured for 8 months before dosing of Fe(II) (5.15 w/w% ferrous sulphate (Dankalk, Løgstør, Denmark)) and Fe(III) (14 w/w% ferric chloride (Brenntag Nordic A/S, Ballerup, Denmark)) was initiated. Fe(II) and Fe(III) were added to the force mains in equal sulphide to iron ratios. Biofilms were adapted to dosing of iron chemicals two months before sampling of biofilm. Samples were taken at 0, 100 and 200 m by scraping with a rubber spatula. The sampling distance of 0 m was located downstream of the chemical injection point at a distance where full mixing of chemicals was ensured. Following collection, the biofilm was centrifuged to remove excess wastewater. DNA/RNA ShieldTM (Zymo Research, Irvine California, US) was added according to manufacturer's guidelines, and samples vortexed to ensure proper conservation. Biofilm samples were kept at −18°C until analysis.
2.2. Microbiome analysis
DNA was extracted from each sample in triplicates using the Quick-DNATM Fecal/Soil Microbe Microprep kit (Zymo Research, Irvine California, US) and 16S rDNA V3+ V4 regions were sequenced on an Illumina MiSeq. Briefly, the V3+ V4 regions were amplified and tagged for sequencing in a two step-PCR procedure using the 341F and 806R primers and PCRBIO HiFi polymerase (PCR Biosystems Ltd., London, UK). Amplicons were purified using Agencourt AMPure XP beads (Beckman Coulter Genomics, MA, US) after each PCR and quantified using Quant-it PicoGreen (Thermo Fisher Scientific Inc, Waltham, MA, USA), before pooling 10 ng DNA from each sample for multiplex sequencing with 10% PhiX using the MiSeq reagent kit V2, PE250 (Illumina, San Diego, CA, USA).
2.3. Bioinformatics
Raw fastq files demultiplexed from the Illumina Miseq were processed using the Dada2 pipeline (Callahan et al., Citation2016) clustering at 97% identity and using the silva_nr_v128_train_set for taxonomic assignment to OTUs. A Neighbour-Joining phylogenetic tree was built using the phangorn package (Schliep, Citation2011). Downstream analysis was carried out in R version 3.3.2 (R Core Team, Citation2018) in R studio version 1.1.383 (RStudio Team, Citation2016) using the metagenomeSeq v1.16.0 (Paulson, Stine, Bravo, & Pop, Citation2013), phyloSeq v1.19.1 (McMurdie & Holmes, Citation2013), Vegan v.2.4.2 (Oksanen et al., Citation2014), ggcorplot v. 0.1.1 and ggplot2 v.2.2.1 (Wickham, Citation2009) packages. Alpha diversity was calculated as the Shannon index, which is a measure of both richness and evenness. Data were filtered for low-abundant Operational Taxonomic Units (OTUs) by removal of OTUs present in fewer than two of the samples and with a relative abundance across all samples ≤0.005% for all but alpha diversity analysis. The phylogenetic tree was rooted before calculating UniFrac distances.
Denoised sequences from Dada2 were exported to a fasta file, replicating each sequence by the amount in the abundance table. Metagenomes were imputed using PICRUSt v.1.1.3 (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) (Langille et al., Citation2013), following the instructions (https://picrust.github.io/picrust/tutorials/quickstart.html). Sequences were reference matched to Greengenes release 13_5, at 97% identity, using the reverse strand matching as recommended and OTU abundances were normalised with the PICRUSt script normalize_by_copy_number.py. Finally, to investigate the functional potential of the microbiome, the 16S rDNA genomic information was inferred with the script predict_metagenomes.py to get information on KEGG ortholog (KO) abundance.
2.4. Statistical analysis
SigmaPlot 13.0 software was used to perform one-way ANOVA followed by Holm–Sidak adjusted multiple pairwise comparisons on alfa diversity and normalised abundances for significantly different bacteria and archaea, when data displayed equality of variances as judged by Brown–Forsythes test. In case of lack of variance homogeneity and normal distribution, data were log-transformed to conform to the assumptions for performing parametrical tests. R was used to identify significantly different bacteria between two groups using the metagenomeSeq package fitZig wrapper on normalised data. Weighted UniFrac distances, which applies both abundance and phylogenetic relationship between the OTUs, were used to calculate the dissimilarity between groups on OTU level, while Bray distances, which solely takes abundance into consideration, were used on the functional KO level. In both cases, the distance matrices were ordinated for visualisation using Principal Coordinate Analysis (PCoA) and overall differences between groups in PCoA plots were analysed using Adonis test (Permutational Multivariate Analysis of Variance Using Distance Matrices) from the Vegan package in R. In all cases the significance level was set to p < 0.05.
3. Results and discussion
Recent advances in multiplex sequencing have made it possible to get information on culturable and non-culturable prokaryotes in a community at the same time in a fast and cost-effective way. Thus, next-generation sequencing of the prokaryotic 16S rDNA hypervariable regions 3 and 4 (V3+ V4) was employed to describe the longitudinal changes in microbial composition in the force main biofilms. Furthermore, the effect of two different iron treatments for sulphide abatement on the biofilm microbiomes was investigated.
3.1. Longitudinally changes in an untreated force main biofilm microbiome
The dominant prokaryotic phyla in the Untreated control main were Firmicutes, Synergistetes, and Proteobacteria, which together constituted approx. 80% of the phyla present. At the genus level, the most dominant genera were Cloacibacillus, Anaerovorax, Fusibacter, Arcobacter, Acetobacterium, Tissierella, and Aminomonas (Figure ).
Figure 1. Taxa summary plot showing spatially differences in abundance of the Untreated main biofilms for the different phyla (a) and genera (b) in the Untreated main biofilm at 0, 100 and 200 m. At the genus level, only the 20 most abundant genera are shown for simplicity. OTUs which could not be assigned to some known genera (categorised as no_match) were omitted. (c) Alpha diversity calculated as the Shannon Index. Values with no capital letters in common are significantly different (p < 0.05). (d) Principal Coordinate Analysis with Weighted UniFrac distances. The 15 most diverse prokaryotes between the three distances were added as vectors pointing in the direction of increased abundance.
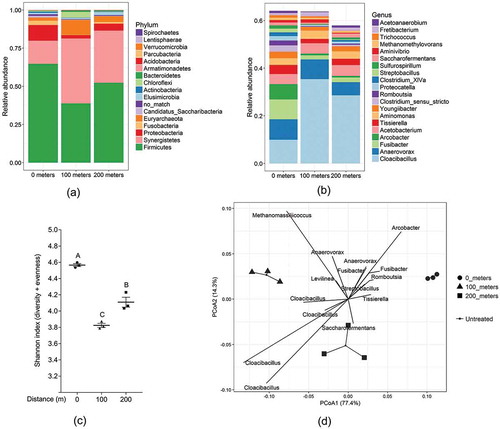
The microbiome composition was observed to change throughout the force main when comparing the abundance profiles of the most abundant OTUs (Figure ). However, a wide range of both high and low abundant prokaryotes were found to be significantly altered throughout the main (see supplementary excel file tab #1). Furthermore, the species diversity (alpha diversity, calculated as the Shannon index, which is a measure of the number of different OTUs and their evenness) changed along the main (p = 0.0035 by one-way ANOVA) (Figure ). Specifically, the species diversity decreases at 100 m compared to 0 m followed by an increase at 200 m. Additionally, changes in global microbiome composition and phylogeny were further confirmed when calculating the dissimilarity index using Weighted UniFrac distances. This method takes both the abundance and phylogenetic relationship into account when determining the difference in microbiome composition between samples. When plotted in a PCoA plot, a significant separation of the microbiome composition at 0, 100 and 200 m can be observed as determined by Adonis test (p < 0.001) (Figure ).
The microbiome composition in the Untreated main was hence observed to change both in species diversity and taxa distribution throughout the main, which shows a highly dynamic prokaryotic community in the force main biofilm. This is in good agreement with previous findings of (Mohanakrishnan et al., Citation2009) who hypothesised changes in microbial populations to be a consequence of variability in types and availability of substrates along the length of the main. This is possible as substrates previously have been shown to exhibit spatial variability in force mains by (Rudelle, Nielsen, Hvitved-Jacobsen, Jensen, & Vollertsen, Citation2016).
3.2. Influence of Fe(II) and Fe(III) treatment on biofilm microbiome
Similar to the observation in the Untreated main (Figure ), longitudinal variations were also observed in the two iron-treated mains, where the abundance of phyla and genus level changed according to sampling distance. At 0 m, dosing of Fe(III) induced larger changes in the microbiome composition of the biofilm compared to Fe(II), with a decrease in Firmicutes and an increase in Synergistetes and Proteobacteria (Figure ). On the genus level, increase in the abundance of Cloacibacillus, Aminomonas and Sulfurospirillum in the Fe(III)-treated main compared to the Untreated and Fe(II)-treated mains were the most apparent changes (Figure ).
Figure 2. Taxa summary plots at phylum level sorted according to the Untreated (UT) main. (a) 0 m; (d) 100 m; (g) 200 m. Taxa summary plots at the genus level, sorted according to the 20 most abundant genera in the Untreated main at the specific distance. (b) 0 m; (e) 100 m; (h) 200 m. OTUs at the genus level which could not be assigned to some genera are omitted (categorised as no_match). Shannon diversity index at (c) 0 m; (f) 100 m; (i) 200 m. Values with no letters in common are significantly different.
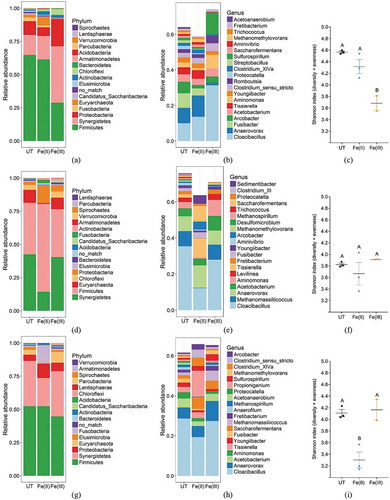
Significant decreases in species diversity, calculated as the Shannon index, due to Fe(III) and Fe(II) dosing were observed at 0 m (p = 0.0044 by one-way ANOVA) and 200 m (p = 0.0063), respectively (Figure and )). Generally, decreased diversity indicates a less complex microbiome in the biofilm which might decrease multifunctionality of the microbial community (Peter et al., Citation2011). This shift in diversity could be due to a selective pressure put on the biofilms by the iron dosing, which in biofilms used for decontamination of mercury-polluted chlor-alkali wastewater similarly was observed to decrease diversity (von Canstein, Kelly, Li, & Wagner-Döbler, Citation2002).
At 100 and 200 m, Fe(II) dosing induced most changes to the biofilm microbiome compositions. At 100 m, a decrease in Synergistetes and Euryarchaeota and an increase of Firmicutes and Proteobacteria on the phylum level was observed, compared to the Untreated and the Fe(III) main (Figure )). Sulphate- and sulphur-reducing bacteria were mainly found in the class of the Deltaproteobacteria, which belongs to the Proteobacteria phylum. The observed increase of the Proteobacteria suggest that genera within this phylum thrived with Fe(II) dosing. The archaeal phylum of Euryarchaeota contains among others the methanogenic archaea, which might suggest a decrease in the methanogenic population in the Fe(II) main. This is also reflected on the genus level where Methanomassiliicoccus were observed to decrease in abundance in parallel with a decrease in Cloacibacillus and increase in, Tissierella, Saccharofermentans, and Sedimentibacter (Figure ).
This pattern was also seen at 200 m, where the abundance of Euryarchaeota decreased while Proteobacteria, as well as Fusobacteria abundances increased in the Fe(II) main (Figure ). At the genus level, Fe(II) dosing increased the genera of Saccharofermentans, Propionigenium, Clostridium sensu stricto, and Arcobacter, with a simultaneous decrease in Cloacibacillus (Figure ). In the Fe(III) main an increase in the genus of Methanomassiliicoccus was the most pronounced change compared to both the Untreated and the Fe(II)-treated mains.
Differences in microbiome composition were further explored using Weighted UniFrac distances of OTU abundances ordinated onto PCoA plots (Figure ). Additionally, differences at the functional level of the microbiome were analysed using PICRUSt, where genomic information from the 16S rDNA sequencing was used to predict the functional potential of the microbiome, i.e. presence and abundance of prokaryotic proteins, by mapping to the KEGG database. Subsequently, relative abundances of the identified KEGG orthologs (KOs) were analysed using Bray distances in PCoA plots. A significant separation in microbiome composition due to iron dosing were observed on both the OTU (0 m p = 0.005, 100 m p = 0.011 and 200 m p = 0.0033) and KO (0 m p = 0.002, 100 m p = 0.009 and 200 m p = 0.005) level at all positions for both iron treatments (Figure ).
Figure 3. PCoA plot with Weighted UniFrac distances showing separation of the microbiome composition at the three positions (a) 0 m; (b) 100 m; (c) 200 m. PCoA plots with Bray distances of relative abundance of KEGG orthologs (KOs) at 0 m (d), 100 m (e), and 200 m (f). Individual prokaryotic and KO abundances were projected onto the ordination and displayed as vectors pointing in the direction of increased abundance. Only the 15 most extreme prokaryotic and KO vectors are shown for simplicity.
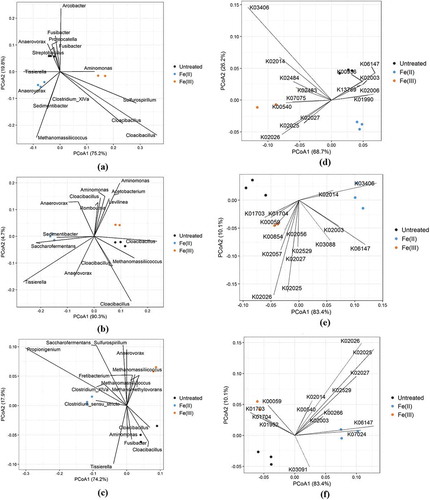
At 0 m Methanomassiliicoccus and Sedimentibacter were driving the separation of the Fe(II) cluster, whereas Sulfurospirillum and Cloacibacillus were driving the Fe(III) cluster away from the Untreated cluster. On the functional level, KOs representing several metabolic pathways (see supplementary excel file tab #5) were more abundant in both the Fe(II) and the Fe(III) samples compared to the Untreated samples.
At 100 m Sedimentibacter still drove the separation of the Fe(II) cluster, but at this distance together with Saccharofermentans and Tissierella. Separation of the Fe(III) cluster from the Untreated cluster was mainly driven by Aminomonas, Acetobacterium, and Cloacibacillus. On the functional level KOs representing metabolic pathways were abundantly driving the separation of the Fe(III) cluster, while KOs belonging to environment sensing and signal transduction were more abundant in the Fe(II) cluster than the other two clusters.
The fermentative Propionigenium genus was mainly responsible for driving the separation of the Fe(II) cluster at 200 m. Whereas separation of the Fe(III) cluster at this distance was driven by the methanogenic Methanomassiliicoccus and Methanomethylovorans together with the genus of Sulfurospirillum. Again, the Fe(III) cluster exhibited increased abundance of KOs related to metabolism, while the Fe(II) cluster had increased abundance of both metabolic and environment sensing KOs.
Taken together, iron dosing in sewer force mains had a significant impact on the biofilm microbiome composition at all positions. This was observed for species diversity, on various phylogenetic levels and similarly on the functional level (KO). Between the 20 most abundant prokaryotic genera (Figures and ) only a few bacteria related to sulphide production (Desulfomicrobium and Sulfurospirillum) and archaea related to methane production (Methanospirillum, Methanomassiliicoccus, and Methanomethylovorans) were observed. Thus, to investigate the influence on prokaryotes related to these processes, specific genera were selected for further examination.
3.3. Effects on genera related to sulphide and methane production
Critical issues during transport of wastewater in force mains relate to sulphide and methane production. Therefore, normalised abundance data specifically for bacteria and archaea related to sulphide and methane production based on literature search and nomenclatural prefixes (desulfo, sulfu and methano) were extracted. Differences in abundance of these prokaryotes between treatments are shown for all three positions in Figure .
Figure 4. Differences in abundance of genera related to sulphide (left) and methane (right) production due to iron dosing at all three positions. Normalised abundance data for genera with the prefix desulfo, sulfu or methano were extracted from the whole dataset for plotting in a bar graph. Bars, within one genus, with no capital letters (A, B, C) in common are significantly different. Note the logarithmic y-axis scale.
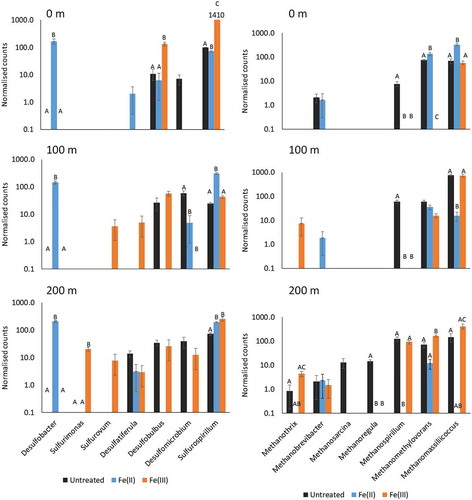
From the sulphide producing genera, Desulfobacter was abundant solely in the Fe(II)-treated main at all distances. Desulfobulbus had a significant higher abundance at 0 m in the Fe(III)-treated main, whereas at the other distances no significant differences between the mains were observed. Sulfurospirillum was highly abundant in all three mains. At 0 m Sulfurospirillum was upregulated in the Fe(III)-treated main compared to both the Untreated and Fe(II)-treated mains. At 100 m the increase in abundance was shifted to the Fe(II)-treated main, and at 200 m both iron-treated mains had an increase of Sulfurospirillum compared to the Untreated main. different species of Sulfurospirillum are known to utilise electron acceptors such as ferric iron, nitrate and elemental sulphur for respiration (Garrity, Citation2005), which all potential could be present in the wastewater or biofilms.
For the genera related to sulphide, there were distinct differences between the Fe(II) and Fe(III)- treated mains even though sulphide ultimately was precipitated to ferrous sulphide in both mains. For some of the genera in the Fe(III)-treated main, elemental sulphur can be used either as electron acceptor or electron donor, which correlates with its possible presence as a product in the reduction of ferric iron as demonstrated by (Davydov, Chuang, & Sanger, Citation1998) using FTIR spectroscopy. Desulfobacter was not identified in this main, which could be because elemental sulphur has been shown to inhibit growth of Desulfobacter (Garrity, Citation2005). However, why Desulfobacter was upregulated in the Fe(II)-treated main compared to the Untreated main, where elemental sulphur was not expected to be present in large amounts, is unknown. It could possibly be a result of the significant differences in the populations of fermenters and other prokaryotes observed in the mains. Thus, changes in the microbiome due to changes in, for example, metabolite availability may have affected other parts of the prokaryotic network and thereby the abundance of sulphate reducing bacteria (Fernandez et al., Citation1999). Additionally, among the 20 most abundant taxa only Sulfurospirillum and Desulfomicrobium were present, and fermenters must, therefore, be expected to have contributed more to the overall decomposition of organic matter and given rise to different fermentation products. Where Desulfobacter primarily utilises acetate as substrate, the other genera related to sulphide production exhibit more diversity in substrate preferences (Garrity, Citation2005).
Regarding the methanogens, Methanomethylovorans and Methanomassiliicoccus showed increased abundance in the Fe(II)-treated main at 0 m. At 100 m no significant difference was observed for Methanomethylovorans between the mains, but at 200 m the Fe(III)-treated main had a higher abundance of Methanomethylovorans than either of the two other mains. Methanomassiliicoccus exhibited a decreased abundance at 100 m in the Fe(II)-treated main, whereas at 200 m this main did not exhibit a significant difference from the Untreated main. Methanospirillum was generally downregulated in both iron-treated mains at all distances compared to the Untreated main.
3.4. Interactions between sulphide and methane related genera
The competition between sulphate-reducers and methanogens for substrates might explain the difference in presence of methanogens observed between the mains. Sulphide and methane production has been shown to proceed simultaneously in anaerobic digestion as well as in sewer sediments (Isa, Grusenmeyer, & Verstraete, Citation1986; Liu, Ni, Ganigué, Werner, Sharma, & Yuan, Citation2015). In the presence of sulphate, sulphate-reducing bacteria in an anaerobic fixed-bed biofilm reactor have been shown to dominate compared to methanogens (Raskin & Rittmann, Citation1996) and the difference in substrate affinity has been demonstrated to account for this, in a competition between Desulfovibrio vulgaris and Methanobrevibacter arboriphilus (Kristjansson, Schönheit, & Thauer, Citation1982). This implies that the methanogens are a result of the sulphate-reducing population. In the Fe(II)-treated main Desulfobacter had a high abundance. Desulfobacter species perform complete oxidation of acetate to CO2 (Castro, Williams, & Ogram, Citation2000; Garrity, Citation2005). The methanogens mainly present in this main, Methanomassiliicoccus and Methanomethylovorans, have preference for H2/CO2, methanol and methylated sulphides for methane production (Cha et al., Citation2013; Dridi, Fardeau, Ollivier, Raoult, & Drancourt, Citation2012; Lomans et al., Citation1999). In the Untreated and Fe(III)- treated main, sulphate-reducers performed incomplete oxidation (Castro et al., Citation2000), leaving formate for the methanogens, which correlates with the presence of e.g. Methanospirillum and Methanoregula in these mains, archaea that for some species use formate and H2/CO2 for methane production (Rosenberg, Citation2014). In marine sediments under high-sulphate conditions, acetate consumption is linked to Desulfobacter. At low-sulphate conditions in contrary, Methanosarcinales takes over in utilising the available acetate for methane production (Purdy, Munson, Cresswell-Maynard, Nedwell, & Embley, Citation2003). However, which specific genera within the order of Methanosarcinales responsible was not identified. Methanosarcina and Methanothrix both belong to this order. These were not identified in the Fe(II)-treated main where Desulfobacter dominated, whereas Methanomethylovorans also belonging to the order had a high abundance, but as discussed earlier this genus has complementary substrate preference to Desulfobacter.
3.5. Ferric iron reducing genera
Besides Sulfurospirillum, no other genera able to conserve energy from ferric reduction according to the table of Dworkin et al. (Citation2006), was detected in the microbiome. This could be due to the proposed difficulty in separating these genera using partial 16s rDNA sequencing as suggested by Lonergan et al. (Citation1996). However, the lack of ferric iron reducers could be a consequence of the chemical state of the iron. The acidic ferric iron added to the main would under near neutral pH conditions rapidly form amorphous or microcrystalline ferric (oxy)hydroxides (Cooper, Picardal, Schimmelmann, & Coby, Citation2003; Davydov et al., Citation1998), where an increased crystallinity will make it less available for bacterial respiration (Lovley & Phillips, Citation1986). However, this state of the ferric iron is still available for sulphide precipitation, when sulphide is formed (dos Santos Afonso & Stumm, Citation1992), with the reactivity governed by crystallinity (Poulton, Krom,, & Raiswell, Citation2004). Fe(III) could furthermore have been used in other reactions in the wastewater, such as phosphate and organic matter precipitation (Metcalf and Eddy, Citation2003), which also would decrease the amount of ferric iron available for respiration.
The absence of ferric reducers could also be a consequence of a lack of competition for common electron donors among the microorganisms. In sewer force mains electron donors are in excess (Zhang et al., Citation2009) and sulphate non-limiting (Hvitved-Jacobsen et al., Citation2013), this implies that ferric iron reducers in this environment do not have a competitive advantage. As the sulphide producers are not inhibited in the force main by this competition, sulphide production would be able to proceed (Lovley & Phillips, Citation1987). Judged visually on the blackening of the wastewater and biofilms during sampling, ferrous sulphides build-up took place in the Fe(III)-treated main. The only genus capable of conserving energy for growth by ferric iron reduction identified was Sulfurospirillum, which furthermore can use other electron acceptors. The increase in redox potential caused by addition of ferric iron to anaerobic wastewater as demonstrated in (Nielsen, Lens, Vollertsen, & Hvitved-Jacobsen, Citation2005), seems unexploited by ferric iron reducers to an extent which would eliminate sulphate reduction and methane production. This implies that sulphate-reducing and methane-producing bacteria in the force main were not outcompeted and hence production of sulphide and methane could proceed during ferric iron treatment.
3.6. Differences on the functional level
Even though the Fe(II) and Fe(III) treatment induced significant alterations to the microbiome community on the genus level compared to the Untreated main, this does not imply that changes were reflected directly on the functional level. In mixed species populations a functional redundancy, which is able to maintain the overall ecosystem performance despite disturbances, can be experienced (Briones & Raskin, Citation2003; Fernandez et al., Citation1999). Thus, PICRUSt was used as a bioinformatic tool to predict the functional potential of the microbiome, to investigate how iron treatment may affect sulphide and methane metabolism by the biofilm. All identified KOs in the dataset were mapped onto KEGG Pathway reference maps for sulphide and methane metabolism to identify significant differences due to iron treatment. At the functional level, KOs belonging to the sulphur and methane metabolism pathways were observed to differ according to treatment and at the different positions throughout the mains (Figure ) (see the description of individual KOs in supplementary excel file tab #6)
Figure 5. Differences in the functional potential of the microbiome related to sulphur and methane metabolism. Bars, within one function, with no capital letters (A, B, C) in common are significantly different.
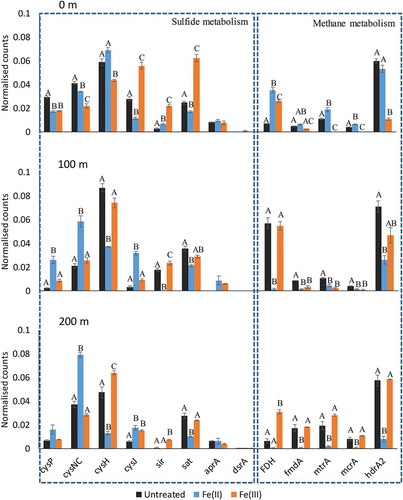
For sulphur metabolism, cysP (K02048), which is part of the cysPUWA sulphate-transporting ATPase responsible for transporting sulphate into the prokaryotic cells, exhibited an increase in the Fe(II) main at 100 m compared to both the Untreated and Fe(III) mains, whereas at 0 m both iron-treated mains exhibited a decrease. The sulphate adenylyltransferase, sat (K00958), represents the first step in both the assimilatory and the dissimilatory sulphate reduction pathways. Here an overall consistent decrease of sat in the Fe(II) main was observed at all distances. This implies that there was a decreased sulphate reduction proceeding in the Fe(II) main. However, abundance of the other proteins involved in dissimilatory sulphate reduction (aprA (K00394) and dsrA (K11180)) were not observed to be significantly affected between the treatments. In the assimilatory sulphate reduction pathway, the sulphite reductase enzyme, sir (K00392), exhibited a consistently increased abundance in the Fe(III) main compared to the other mains, indicating an increased sulphide accumulation.
In the Fe(II) and to a lesser extent the Fe(III)-treated main, at 100 and 200 m, a decrease in methanogenic KOs, mtrA (K00577), mcrA (K00399) and hdrA2 (K03388) were observed. This suggests that methane production was affected by iron treatment and that the production might have been decreased compared to the Untreated main. At 100 and 200 m an overall decrease in KOs related to formate metabolism, FDH (K00122) and fmdA (K00200), were observed in the Fe(II) main. This corresponds well with the presence of Desulfobacter that performs complete oxidation of acetate to CO2, as well as with the observed methanogens which, do not have a preference for formate as described previously.
Thus, the microbiome analysis suggests a decrease in the functional potential for sulphate reduction and methane production in biofilms due to iron dosing. This is in good agreement with the measured decrease in sulphate reduction and methane production previously reported for biofilms from the same force main (Kiilerich et al., Citation2018). In these two studies, the addition of Fe(II) and Fe(III) both exhibited comparable effects, indicating that it is not directly due to the increased redox potential of Fe(III) addition. This is further supported by Zhang et al. (Citation2009) who proposed a similar explanation based on observations of inhibition of sulphate reduction and methane production in a reactor supplemented with Fe(III).
4. Conclusions
The current study was performed to investigate the effect of Fe(II) and Fe(III) addition on biofilms microbiomes related to sulphide and methane production in sewer force mains.
Both Fe(II) and Fe(III) dosing caused overall significant changes to the entire prokaryotic community composition and its functional potential, compared to biofilms in an untreated control force main. Changes, which appear dynamic since both treatment and distance from the dosing point differentially affected the microbiomes compositions.
Changes in sulphate reducing bacteria due to iron dosing were also quite complex, apart from the consistent high abundance of Desulfobacter after Fe(II) dosing. Similarly, the functional pathway for dissimilatory sulphate reduction, indicative for sulphide production in the mains, was not unequivocally affected by the iron treatments. Since a decrease in sulphide production due to iron dosing has previously been observed in the same force main, this suggests a broad functional redundancy of the microbiome related to sulphide production.
Lastly, a concurrent reduction in the archaeal methanogenic genera as well as their functional potential after iron dosing, indicative of a reduction of methane production was observed in this study. Furthermore, the distribution of the methanogenic population was closely linked to sulphate reducers present in the specific main. This was most distinctly noticed in the Fe(II)-treated main, where the abundance of methanogenic genera and KOs related to utilisation of formate were lacking.
Author Contributions
B.K., J.V. and P.K. conceived and designed the experiments; B.K. and P.K. performed the experiments; B.K., P.K., A.D.B. analysed the data; B.K. and P.K. wrote the paper, A.D.B and J.V. revised the paper.
Conflicts of Interest
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Correction
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Supplemental Material
Download MS Excel (74 KB)Acknowledgements
This work is partly funded by the Innovation Fund Denmark (IFD) under File No. 4135-00076B.
Supplementary material
Supplementary material for this article can be accessed here.
Additional information
Funding
Notes on contributors
Bruno Kiilerich
Bruno Kiilerich is a senior water treatment specialist at Grundfos Holding A/S with a research interest in hydrogen sulphide abatement. The research is centred around developing dosing algorithms for chemical abatement, with the aim of optimizing the amount of chemicals needed to perform proper abatement. It is therefore of prime necessity to understand the microbial community colonizing sewer systems and how the community responds to treatment.
References
- Achtnich, C., Bak, F., & Conrad, R. (1995). Competition for electron donors among nitrate reducers, ferric iron reducers, sulfate reducers, and methanogens in anoxic paddy soil. Biology and Fertility of Soils, 19, 65–15. doi:10.1007/BF00336349
- Apgar, D., & Witherspoon, J. (2007). Minimization of odors and corrosion in collection systems phase 1. Water Environment Research Foundation (WERF). London: IWA Publishing.
- ASCE, 1989. Sulfide in wastewater collection and treatment systems ASCE (American Society of Civil Engineers) Manuals and Reports on Enginering Practice No. 69, New York, USA. New York, NY: American Society of Civil Engineers.
- Boon, A. G. (1995). Septicity in sewers: Causes, consequences and containment. Water Science and Technology, 31, 237–253. doi:10.1016/0273-1223(95)00341-J
- Briones, A., & Raskin, L. (2003). Diversity and dynamics of microbial communities in engineered environments and their implications for process stability. Current Opinion in Biotechnology, 14, 270–276. doi:10.1016/S0958-1669(03)00065-X
- Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., & Holmes, S. P. (2016). DADA2: High-resolution sample inference from illumina amplicon data. Nature Methods, 13, 581–583. doi:10.1038/nmeth.3869
- Castro, H. F., Williams, N. H., & Ogram, A. (2000). Phylogeny of sulfate-reducing bacteria. FEMS Microbiology Ecology, 31, 1–9. doi:10.1016/S0168-6496(99)00071-9
- Cha, I.-T., Min, U.-G., Kim, S.-J., Yim, K. J., Roh, S. W., & Rhee, S.-K. (2013). Methanomethylovorans uponensis sp. nov., a methylotrophic methanogen isolated from wetland sediment. Antonie van Leeuwenhoek, 104, 1005–1012. doi:10.1007/s10482-013-0020-4
- Cooper, D. C., Picardal, F. W., Schimmelmann, A., & Coby, A. J. (2003). Chemical and biological interactions during nitrate and goethite reduction by Shewanella putrefaciens 200. Applied and Environmental Microbiology, 69, 3517–3525. doi:10.1128/AEM.69.6.3517
- Davydov, A., Chuang, K. T., & Sanger, A. R. (1998). Mechanism of H2S oxidation by ferric oxide and hydroxide surfaces. The Journal of Physical Chemistry, 102, 4745–4752. doi:10.1021/jp980361p
- dos Santos Afonso, M., & Stumm, W. (1992). Reductive dissolution of iron(III) (Hydr)oxides by hydrogen sulfide. Langmuir, 8, 1671–1675. doi:10.1021/la00042a030
- Dridi, B., Fardeau, M.-L., Ollivier, B., Raoult, D., & Drancourt, M. (2012). Methanomassiliicoccus luminyensis gen. nov., sp. nov., a methanogenic archaeon isolated from human faeces. International Journal of Systematic and Evolutionary Microbiology, 62, 1902–1907. doi:10.1099/ijs.0.033712-0
- Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., & Stackebrandt, E. (2006). The prokaryotes, a handbook on the biology of bacteria volume 2: Ecophysiology and biochemistry, third edit. ed, the prokaryotes. New York: Springer Science+Business Media, LLC. doi:10.1007/0-387-30741-9
- Fernandez, A., Huang, S., Seston, S., Xing, J., Hickey, R., Criddle, C., & Tiedje, J. (1999). How stable is stable? Function versus community composition. Applied and Environmental Microbiology, 65, 3697–3704.
- Ganigue, R., Gutierrez, O., Rootsey, R., & Yuan, Z. (2011). Chemical dosing for sulfide control in Australia: An industry survey. Water Research, 45, 6564–6574. doi:10.1016/j.watres.2011.09.054
- Garrity, G. M. (2005). Bergey’s manual of systematic bacteriology - volume two: The proteobacteria (part C). 2nd ed. Boston, MA: Springer-Verlag US. doi:10.1007/0-387-29298-5
- Guisasola, A., de Haas, D., Keller, J., & Yuan, Z. (2008). Methane formation in sewer systems. Water Research, 42, 1421–1430. doi:10.1016/j.watres.2007.10.014
- Hvitved-Jacobsen, T., Vollertsen, J., & Nielsen, A. H. (2013). Sewer processes - microbial and chemical process engineering of sewer networks. Boca Raton, FL, USA: CRC press.
- Isa, Z., Grusenmeyer, S., & Verstraete, W. (1986). Sulfate reduction relative to methane production in high-rate anaerobic digestion: microbiological aspects. Applied and Environmental Microbiology, 51, 580–587.
- Jensen, H. S., Sekar, R., Shepherd, W. J., Osborn, A. M., Tait, S., & Biggs, C. A. (2016). Spatial and temporal variability of bacterial communities within a combined sewer system. Microbiologyopen, 5, 616–625. doi:10.1002/mbo3.356
- Kiilerich, B., Kiilerich, P., Nielsen, A. H., & Vollertsen, J. (2018). Variations in activities of sewer biofilms due to ferrous and ferric iron dosing. Water Science and Technology : a Journal of the International Association on Water Pollution Research, 2017, 845–858. doi:10.2166/wst.2018.261
- Kristjansson, J. K., Schönheit, P., & Thauer, R. K. (1982). Different Ks values for hydrogen of methanogenic bacteria and sulfate reducing bacteria: An explanation for the apparent inhibition of methanogenesis by sulfate. Archives of Microbiology, 131, 278–282. doi:10.1007/BF00405893
- Langille, M. G. I., Zaneveld, J., Caporaso, J. G., McDonald, D., Knights, D., Reyes, J. A., … Huttenhower, C. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotechnology, 31, 814–821. doi:10.1038/nbt.2676
- Liu, Y., Ni, B.-J., Ganigué, R., Werner, U., Sharma, K. R., & Yuan, Z. (2015). Sulfide and methane production in sewer sediments. Water Research, 70, 350–359. doi:10.1016/j.watres.2014.12.019
- Lomans, B. P., Maas, R., Luderer, R., Op Den Camp, H. J. M., Pol, A., van der Drift, C., & Vogels, G. D. (1999). Isolation and characterization of methanomethylovorans hollandica gen. nov., sp. nov., isolated from freshwater sediment, a methylotrophic methanogen able to grow on dimethyl sulfide and methanethiol. Applied and Envirnmental Microbiology, 65, 3641–3650.
- Lonergan, D. J., Jenter, H. L., Coates, J. D., Phillips, E. J. P., Schmidt, T. M., & Lovley, D. R. (1996). Phylogenetic analysis of dissimilatory Fe(III)-reducing bacteria. Journal of Bacteriology, 178, 2402–2408. doi:10.1128/jb.178.8.2402-2408.1996
- Lovley, D. R. (1991). Dissimilatory Fe(III) and Mn(IV) reduction. Microbiology and Molecular Biology Review, 55, 259–287
- Lovley, D. R. (1993). Dissimilatory metal reduction. Annual Reviews in Microbiology, 47, 263–290. doi:10.1146/annurev.mi.47.100193.001403
- Lovley, D. R., & Phillips, E. J. P. (1986). Organic matter mineralization with reduction of ferric iron in anaerobic sediments. Applied and Envirnmental Microbiology, 51, 683–689. doi:10.1080/01490458709385975
- Lovley, D. R., & Phillips, E. J. P. (1987). Competitive mechanisms for inhibition of sulfate reduction and methane production in the zone of ferric iron reduction in sediments. Applied and Envirnmental Microbiology, 53, 2636–2641. doi:10.1016/j.ocecoaman.2011.09.006
- Lovley, D. R., Roden, E. E., Phillips, E. J., & Woodward, J. (1993). Enzymatic iron and uranium reduction by sulfate-reducing bacteria. Marine Geology, 113, 41–53. doi:10.1016/0025-3227(93)90148-O
- McMurdie, P. J., & Holmes, S. (2013). phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One, 8. doi:10.1371/journal.pone.0061217
- Metcalf and Eddy. (2003). Wastewater engineering: Treatment and reuse (4th ed ed.). Boston: McGraw-Hill.
- Mohanakrishnan, J., Sharma, K. R., Meyer, R. L., Hamilton, G., Keller, J., & Yuan, Z. (2009). Variation in biofilm structure and activity along the length of a rising main sewer. Water Environment Research, 81, 800–808. doi:10.2175/106143008X390771
- Nielsen, A. H., Lens, P., Vollertsen, J., & Hvitved-Jacobsen, T. (2005). Sulfide-iron interactions in domestic wastewater from a gravity sewer. Water Research, 39, 2747–2755. doi:10.1016/j.watres.2005.04.048
- Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O’Hara, R. B., … Wagner, H. (2014). Vegan: Community ecology package. R package version 2.2-0 (pp. 10). Retrieved from http://CRAN.Rproject.org/package=vegan
- Paulson, J. N., Stine, O. C., Bravo, H. C., & Pop, M. (2013). Differential abundance analysis for microbial marker-gene surveys. Nature Methods, 10, 1200–1202. doi:10.1038/nmeth.2658
- Peter, H., Ylla, I., Gudasz, C., Romaní, A. M., Sabater, S., & Tranvik, L. J. (2011). Multifunctionality and diversity in bacterial biofilms. PloS one, 6, e23225. doi:10.1371/journal.pone.0023225
- Poulton, S. W., Krom,, M. D., & Raiswell, R. (2004). A revised scheme for the reactivity of iron (oxyhydr)oxide minerals towards dissolved sulfide. Geochimica et cosmochimica acta, 68, 3703–3715. doi:10.1016/j.gca.2004.03.012
- Purdy, K. J., Munson, M. A., Cresswell-Maynard, T., Nedwell, D. B., & Embley, T. M. (2003). Use of 16S rRNA-targeted oligonucleotide probes to investigate function and phylogeny of sulphate-reducing bacteria and methanogenic archaea in a UK estuary. FEMS Microbiology Ecology, 44, 361–371. doi:10.1016/S0168-6496(03)00078-3
- R Core Team. (2018). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. Retrieved from https://www.R-project.org
- Raskin, L., & Rittmann, B. E. (1996). Competition and coexistence of sulfate-reducing and methanogenic populations in anaerobic biofilms. Applied and Environmental Microbiology, 62, 3847–3857.
- Rosenberg, E. (2014). The prokaryotes - other major lineages of bacteria and the archaea (4th ed.). Berlin, Heidelberg: Springer-Verlag. doi:10.1007/978-3-642-38954-2
- RStudio Team. (2016). RStudio: Integrated development for R. Boston, MA: RStudio, Inc. Retrieved from http://www.rstudio.com
- Rudelle, E. A., Nielsen, A. H., Hvitved-Jacobsen, T., Jensen, H. S., & Vollertsen, J. (2016). Spatial variability of anaerobic processes and wastewater pH in force mains. Water Environment Research : a Research Publication of the Water Environment Federation, 88, 747–755. doi:10.2175/106143016X14609975747126
- Schliep, K. P. (2011). phangorn: Phylogenetic analysis in R. Bioinformatics, 27, 592–593. doi:10.1093/bioinformatics/btq706
- von Canstein, H., Kelly, S., Li, Y., & Wagner-Döbler, I. (2002). Species diversity improves the efficiency of mercury-reducing biofilms under changing environmental conditions. Applied and Environmental Microbiology, 68, 2829–2837. doi:10.1128/AEM.68.6.2829
- Wickham, H. (2009). ggplot2 - elegant graphics for data analysis (1st ed.). New York: Springer-Verlag. doi:10.1007/978-0-387-98141-3
- Zhang, L., Keller, J., & Yuan, Z. (2009). Inhibition of sulfate-reducing and methanogenic activities of anaerobic sewer biofilms by ferric iron dosing. Water Research, 43, 4123–4132. doi:10.1016/j.watres.2009.06.013
