Abstract
The complete mitochondrial genome of the koi carp (Cyprinus carpio, Cyprinidae) was sequenced in the present study by using Ion Torrent Personal Genome Machine (PGM) platform for the first time. The mitochondrial genome sequence is 16 581 bp in size and consists of 13 protein-coding genes, 22 tRNA genes, two rRNA genes and one control region. The gene order and organization were similar to most of the other teleost. The nucleotide compositions of the light strand are 24.82% of A, 31.92% of T, 27.53% of G and 15.73% of C. With the exception of eight tRNA genes and the NADH dehydrogenase subunit 6 (ND6), all other mitochondrial genes are encoded on the heavy strand. The phylogenetic tree constructed using a maximum-likelihood model showed sister relationship of koi carp to other Cyprinidae fishes.
The koi carp (Cyprinus carpio, Cyprinidae) include the colourful ornamental varieties of common carp which are represented in various competitive exhibitions worldwide and are probably the most expensive market of individual freshwater fish (FAO Citation2012). To date, all the published mitochondrial genomes of Cyprinus carpio were sequenced by using Illumina Platform (Wang et al. Citation2013; Hu et al., Citation2014a, Citation2014b; Lin et al. Citation2014; Mabuchi & Song Citation2014). Comparison research of Illumina Platform and Ion Torrent PGM platform were indicated that there are key differences between the quality of that data and the applications it will support (Quail et al. Citation2012; Salipante et al. Citation2014). The Ion Torrent Platform was proved could assemble the mitogenome of fish species effectively (Xie et al. Citation2014; Raman et al. Citation2015). Thus, the Ion Torrent Platform was employed in this study aimed at enhancing the accuracy of Cyprinus carpio mitogenome and providing the basis for further study on conservation researches.
The specimen was sampled from a culture farm in Nansha, Guangdong Province, China. The pectoral fin of the fresh fish was preserved in 95% ethanol and the total genomic DNA was extracted by using the salting-out procedure (Howe et al. Citation1997). The entire sequence of koi carp (Cyprinus carpio) mitochondrial genome (GenBank accession number KU159761) is 16 581 bp in length and contained 38 genes including 13 protein-coding genes, 22 transfer RNA genes (tRNA), two ribosomal RNA genes (12S rRNA and 16S rRNA) and a control region. All genes were encoded on the heavy strand, with only the NADH dehydrogenase subunit 6 (ND6) and eight tRNA genes (Gln, Ala, Asn, Cys, Tyr, Glu, Pro, Ser) were encoded on the light strand. The nucleotide compositions of the both light and heavy strand are 33.12% of A, 33.01% of T, 14.24% of G and 19.63% of C.
All of the protein-coding genes begin with an ATG initiation codon except COX1 which was started with GTG. There were three types of termination codons including TAA for ND1, COX1, ATP6, COX3, ND4L, ND5 and ND6, TAG for ND2, ATP8 and ND3, T for COX2, ND4 and CYTB. The length of the 12S rRNA and 16SrRNA was 955 and 1679 bp, respectively. The 12S and 16S rRNA genes were located between the tRNA-Phe (GAA) and tRNA-Leu (TAA) genes, and separated by the tRNA-Val gene with the same location situation was also found in other vertebrates (Yang et al. Citation2014; Zhao et al. Citation2014). The 22 tRNA genes were ranging from 67 (tRNA-Cys) to 76 bp (tRNA-Leu and tRNA-Lys) in length. All these tRNAs could fold into the typical cloverleaf secondary structure although numerous non-complementary and T–G base pairs exist in the stem regions. The control region (D-loop gene) was 927 bp in length, located between tRNA-Pro (TGG) and tRNA-Phe (GAA) gene. Most genes were either abutted or overlapped.
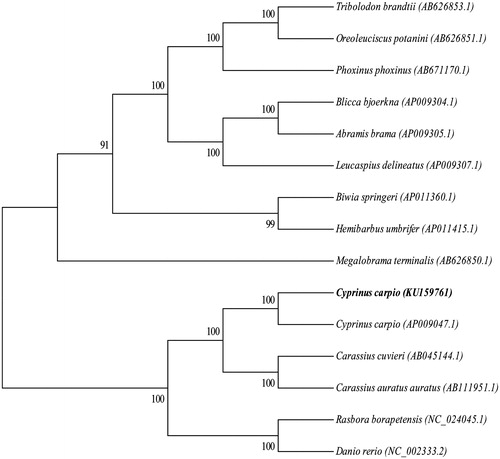
The 15 mitochondrial genomes from GenBank which belongs to the subfamily Cyprinidae were selected to derive phylogenetic relationships (). After analysis by the jModelTest2.1.7, the phylogenetic tree was constructed using RAxML8.1.5 software (Scientific Computing Group, Heidelberg Institute for Theoretical Studies, Heidelberg, Germany) through maximum-likelihood method and the Bootstrap value was 1000 (Stamatakis Citation2006; Posada Citation2008).
Figure 1. Phylogenetic tree generated using the maximum-likelihood method based on complete mitochondrial genomes. GenBank accession numbers for the published sequences are Tribolodon brandtii (AB626853.1), Oreoleuciscus potanini (AB626851.1), Phoxinus phoxinus (AB671170.1), Blicca bjoerkna (AP009304.1), Abramis brama (AP009305.1), Leucaspius delineatus (AP009307.1), Biwia springeri (AP011360.1), Hemibarbus umbrifer (AP011415.1), Megalobrama terminalis (AB626850.1), Cyprinus carpio L. (AP009047.1), Carassius auratus cuvieri (AB045144.1), Carassius auratus auratus (AB111951.1), Rasbora borapetensis (NC_024045.1) and Danio rerio (NC_002333.2)
Disclosure statement
The authors report no conflicts of interest. The authors themselves are responsible for the content and writing of the paper.
Funding information
This research was supported by the National Natural Science Foundation of China under grant No. 31001122; the National Basic Research Program of China under grant No. 2012CB114406; Technology Planning Project of Guangdong Province under number 2011A020102002 and the special supporting project 20090450193 of China Postdoctoral Science Foundation.
References
- FAO. 2012. Statistics and Information Service of the Fisheries and Aquaculture Department: Fishery and Aquaculture Statistics 2010.
- Howe JR, Klimstra DS, Cordon-Cardo C. 1997. DNA extraction from paraffin-embedded tissues using a salting-out procedure: a reliable method for PCR amplification of archival material. Histol Histopathol. 12:595–601.
- Hu GF, Liu XJ, Li Z, Liang HW, Hu SN, Zou GW. 2014a. Complete mitochondrial genome of Xingguo red carp (Cyprinus carpio var. singuonensis) and purse red carp (Cyprinus carpio var. wuyuanensis). Mitochondrial DNA.
- Hu GF, Liu XJ, Zou GW, Li Z, Liang HW, Hu SN. 2014b. Complete mitochondrial genome of Yangtze River wild common carp (Cyprinus carpio haematopterus) and Russian scattered scale mirror carp (Cyprinus carpio carpio). Mitochondrial DNA.
- Lin M, Zou J, Wang C. 2014. Complete mitochondrial genomes of domesticated and wild common carp (Cyprinus carpio L.). Mitochondrial DNA.
- Mabuchi K, Song H. 2014. The complete mitochondrial genome of the Japanese ornamental koi carp (Cyprinus carpio) and its implication for the history of koi. Mitochondrial DNA 25:35–36.
- Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25:1253–1256.
- Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, Bertoni A, Swerdlow HP, Gu Y. 2012. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics 13:341.
- Raman S, Pavan-Kumar A, Koringa PG, Patel N, Shah T, Singh RK, Krishna G, Joshi CG, Gireesh-Babu P, Chaudhari A, et al. 2015. Ion torrent next-generation sequencing reveals the complete mitochondrial genome of endangered mahseer Tor khudree (Sykes, 1839). Mitochondrial DNA.
- Salipante SJ, Kawashima T, Rosenthal C, Hoogestraat DR, Cummings LA, Sengupta DJ, Harkins TT, Cookson BT, Hoffman NG. 2014. Performance comparison of Illumina and ion torrent next-generation sequencing platforms for 16S rRNA-based bacterial community profiling. Appl Environ Microbiol. 80:7583–7591.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690.
- Wang B, Ji P, Wang J, Sun J, Wang C, Xu P, Sun X. 2013. The complete mitochondrial genome of the Oujiang color carp, Cyprinus carpio var. color (Cypriniformes, Cyprinidae). Mitochondrial DNA 24:19–21.
- Xie Z, Yu C, Guo L, Li M, Yong Z, Liu X, Meng Z, Lin H. 2014. Ion Torrent next-generation sequencing reveals the complete mitochondrial genome of black and reddish morphs of the Coral Trout Plectropomus leopardus. Mitochondrial DNA.
- Yang H, Zhao H, Sun J, Chen Y, Liu L, Zhang Y, Liu L. 2014. The complete mitochondrial genome of the Hemibarbus medius (Cypriniformes, Cyprinidae). Mitochondrial DNA.
- Zhao H, Yang H, Sun J, Chen Y, Liu L, Li G, Liu L. 2014. The complete mitochondrial genome of the Anabas testudineus (Perciformes, Anabantidae). Mitochondrial DNA.
