Abstract
Corbicula fluminea is an important aquatic commercial species in China. In this study, we present the complete mitogenome and a phylogenetic analysis of C. fluminea, determined using next-generation and PacBio long read sequencing. The mitogenome of C. fluminea is 17,423 bp in size, including 13 protein-coding genes, two ribosomal RNA genes, 22 tRNA genes, and a putative control region, all located on the same strand. The base composition of the entire mitogenome showed a conspicuous A + T bias of 70.5 %. The entire mitogenome data produced in this study provides the genomic resources available for future evolutionary studies.
Corbicula fluminea (Müller, 1774) is a species of bivalve mollusks. The extant native distribution of this freshwater suspension feeder is Asia, the Middle East, Africa, and Australia (Mcmahon Citation1982). Since the early 20th century, it spreads over the world through commercialization, occupies the benthic sediment in rivers, lakes and estuaries, and becomes an invasive alien species (IAS) to American and European ecosystems (Counts Citation1981; Phelps Citation1994; Mouthon Citation2001; Schmidlin and Baur Citation2007; Sousa et al. Citation2008; Caffrey et al. Citation2011; Beghelli et al. Citation2014; Ilarri et al. Citation2014; Molina et al. Citation2015). As an edible aqua-cultural shellfish, over 12 million tons C. fluminea is output per year (Chen et al. Citation2013), making it to be one of the most important aquatic commercial species in China.
In this study, we characterized the complete mitogenome sequence of C. fluminea using Illumina next-generation and Pacific Biosciences (PacBio) sequencing technology and performed an analysis of the phylogenetic relationships among Veneroida.
We collected the clam (NO. HX_HZ092701) from Hongze Lake (33.2443 N and 118.4566 E) on 27 September 2017. Purified mitochondria were extracted from muscle tissue homogenate by differential centrifugation, and subsequently, DNA was isolated following Sambrook and Russell (Citation2001). The specimen was stored in the herbarium of Freshwater Fisheries Research Institute of Jiangsu Province.
Mitochondrial genome was sequenced and reconstructed based on both Illumina Hiseq (Borgström et al. Citation2011) and the Pacbio Sequel (English et al. Citation2012) data. 5,545,482,944 clean reads were generated and assembly was performed using ABySS (Simpson et al. Citation2009). Genes were annotated using MITOS (Matthias et al. Citation2013) and AUGUSTUS (http://bioinf.uni-greifswald.de/augustus/). tRNA and rRNA genes were predicted using tRNAscan-SE (Lowe and Eddy Citation1997) and rRNAmmer 1.2 (Lagesen et al. Citation2007), respectively.
The mitogenome of C. fluminea (HX_HZ092701) is 17,423 bp long (GenBank accession no. MK392334), has a 29.5% G + C content, and encodes 37 genes, including 13 protein-coding genes (PCGs), two rRNA genes, and 22 tRNA genes. All genes located on the same strand, compared with the majority of bivalves. Most protein-coding genes have ATG as the start codon, while two PCGs (cox1 and nad5) use ATT and nad4L uses ATA. TAA and TAG stop codons are used in most genes, excepted for four genes, nad1-4, which have incomplete stop codon TA.
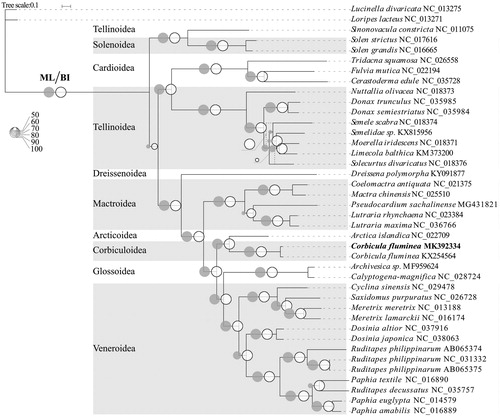
To investigate the phylogenetic relationships of C. fluminea and other 36 species from nine superfamilies in Veneroida, phylogenetic trees were obtained using Maximum-Likelihood (ML) and Bayesian inference (BI) analyses based on entire PCGs sequences. Two species in the order Lucinoida were used as outgroup (). The consensus topology of phylogeny trees for 38 Veneroida species shows that Corbiculoidea and Arcticoidea is a sister group. Each superfamily in Veneroida is monophyly. It is worth noting that a classical barcoding fragment (cox1) shows an obvious genetic divergency between a partial genome of C. fluminea (KX254564) and our sequences. Further investigation will be needed to reveal the reason for this genetic divergency.
Figure 1. Phylogenetic tree in Veneroida using the complete mitochondrial PCGs sequences. The complete mitogenome is downloaded from GenBank and the phylogenic tree is constructed by maximum-likelihood (ML) and Bayesian inference (BI) method. The bootstrap values and posteriori probability were presented as the proportion of gray and white circles, respectively. The dotted lines indicated the structure of the BI tree differing from that of ML tree. The genomic sequences obtained in this study were shown by bold type.
Acknowledgements
The authors would like to thank Yan Cheng and Cun Jing for their helpful suggestions.
Disclosure statement
The authors report that they have no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Beghelli FGDS, Pompêo MLM, Carlos VM. 2014. First occurrence of the exotic Asian clam Corbicula fluminea (Müller, 1774) in the Jundiaí-Mirim River Basin, SP, Brazil. Ambient Água. 9(3):402.
- Borgström E, Lundin S, Lundeberg J. 2011. Large scale library generation for high throughput sequencing. PLoS One. 6(4):e19119.
- Caffrey JM, Evers S, Millane M, Moran H. 2011. Current status of Ireland's newest invasive species – the Asian clam Corbicula fluminea (Müller, 1774). Aquat Invasions. 6:391–399.
- Chen H, Zha J, Liang X, Bu J, Wang M, Wang Z. 2013. Sequencing and de novo assembly of the Asian clam (Corbicula fluminea) transcriptome using the Illumina GAIIx method. PLoS One. 8(11):e79516.
- Counts CL. 1981. Corbicula fluminea (Bivalvia: Sphaeriacea) in British Columbia. Nautilus. 95:12–13.
- English AC, Richards S, Han Y, Wang M, Vee V, Qu J, Qin X, Muzny DM, Reid JG, Worley KC et al. 2012. Mind the gap: upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS One. 7(11):e47768.
- Ilarri MI, Souza AT, Antunes C, Guilhermino L, Sousa R. 2014. Influence of the invasive Asian clam Corbicula fluminea (Bivalvia: Corbiculidae) on estuarine epibenthic assemblages. Estuarine Coastal Shelf Sci. 143(4):12–19.
- Lagesen K, Hallin P, Rodland E, Staerfeldt H, Rognes T, Ussery D. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35(9):3100.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964.
- Matthias B, Alexander D, Frank J, Fabian E, Catherine F, Guido F, Joern P, Martin M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Mcmahon RF. 1982. The occurrence and spread of the introduced Asiatic freshwater clam, Corbicula fluminea (Müller), in North America 1924–1982. Nautilus. 96:134–141.
- Molina LM, Pereyra PJ, Carrizo NGM, Abrameto MA. 2015. Here come the clam: Southernmost record worldwide of the Asian clam Corbicula fluminea (Patagonia, Argentina). Russian J Biol Invasions. 6(2):129–134.
- Mouthon J. 2001. Life cycle and population dynamics of the Asian clam Corbicula fluminea (Bivalvia: Corbiculidae) in the Saone River at Lyon (France). Hydrobiologia. 452(1–3):109–119.
- Phelps HL. 1994. The Asiatic clam (Corbicula fluminea) invasion and system-level ecological change in the Potomac River Estuary near Washington, D.C. Estuaries. 17(3):614–621.
- Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. 3rd ed. New York: Cold Spring Harbor Laboratory.
- Schmidlin S, Baur B. 2007. Distribution and substrate preference of the invasive clam Corbicula fluminea in the river Rhine in the region of Basel (Switzerland, Germany, France). Aquat Sci. 69(1):153–161.
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res. 19:1117–1123.
- Sousa R, Antunes C, Guilhermino L. 2008. Ecology of the invasive Asian clam Corbicula fluminea (Müller, 1774) in aquatic ecosystems: an overview. Ann Limnol. 44(2):85–94.
