Abstract
Aquilaria crassna Pierre ex Lecomte is a principal agarwood tree resource. It has been listed as a critically endangered species. The complete chloroplast genome of A. crassna was assembled based on next-generation sequencing. The plastome was a quadripartite circular shape with 174,830 bp in length, including a large single-copy (LSC) region of 87,450 bp and small single-copy (SSC) region of 3346 bp, separated by two inverted repeat (IR) regions of 42,017 bp each. The chloroplast genome contained 125 genes, including 79 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. The overall GC content of the whole genome was 36.7%. The phylogenetic tree showed that three species of Aquilaria formed one monophyletic clade with 100% bootstrap value.
The genus Aquilaria, including 21 accepted species native to Southeast Asia, is a well-known genus as the principal resource of the resinous agarwood used in incense, perfume, and traditional drug with high value in many cultures (Hashim et al. Citation2016; Zhang et al. Citation2019). Aquilaria crassna Pierre ex Lecomte is a representative agarwood tree mainly distributed in the primary and secondary forest in Cambodia, Vietnam, Thailand, and Laos (Ng et al. Citation1997; Barden et al. Citation2000). Due to overwhelming illegal harvesting for increasing demand of agarwood products, the population of A. crassna decreased rapidly in recent years. Aquilaria crassna has been listed in IUCN Red List of Threatened Species as Critically Endangered species with status ‘A2cd’ (Harvey-Brown Citation2018). It is an emergency to carry out suitable conservation strategy to protect this species. However, fundamental genetic information about this species remains less until now. Here, we reported the complete chloroplast genome of A. crassna to provide genomic resource for further conservation genetics and phylogenetic analysis.
The fresh leaves were collected from a healthy A. crassna tree, introduced from Cambodia, growing in the Xing Long Tropical Medicinal Botanical Garden (18°43′56″N 110°11′43″E) in Hainan Province. The voucher specimens (ZhangHN02) were deposited in the Herbarium of Yunnan Normal University. A sequence library was constructed and sequencing was performed using the Illumina HiSeq 2500-PE150 platform (Illumina, San Diego, CA, USA). All raw reads were filtered using NGS QC Toolkit_v2.3.3 with default parameters to obtain clean reads (Patel and Jain Citation2012). The plastome was de novo assembled using NOVOPlasty (Dierckxsens et al. Citation2017). The complete cp genome was annotated with the online annotation tool GeSeq (Tillich et al. Citation2017). The SSRs were detected using online software IMEx (Mudunuri and Nagarajaram Citation2007).
The complete chloroplast genome of A. crassna (Genbank accession no.: MK779998) was quadripartite circular shape with 174,830 bp in length, including a large single-copy (LSC) region of 87,450 bp and small single-copy (SSC) region of 3346 bp, separated by two inverted repeat (IR) regions of 42,017 bp, respectively. The overall GC content was 36.7%. The chloroplast genome contained 79 protein-coding genes, 38 tRNA genes, and eight rRNA genes. Total 71 simple sequence repeats (SSRs) were discovered in the complete chloroplast genome, of which mono-nucleotide SSR motif was most frequently distributed.
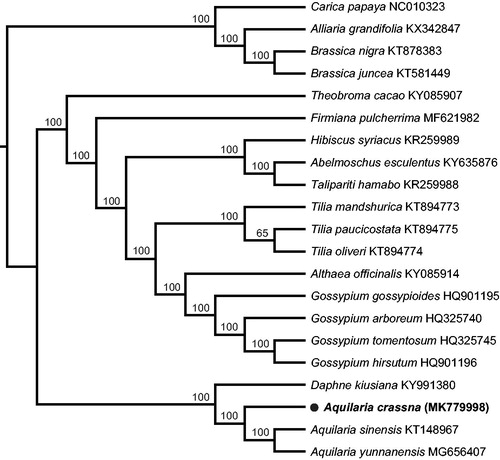
In order to understand the phylogenetic relationship between A. crassna and related species, the published chloroplast genomes, including A. sinensis, A. yunnanensis and Daphne kiusiana from Thymelaeaceae and 13 species from Malvaceae, with four species from Brassicales as outgroups, were aligned by MAFFT 7.308 (Katoh and Standley Citation2013). The maximum likelihood (ML) tree was constructed with RAxML 8.2.11 (Stamatakis et al. Citation2008) and the branch support was estimated with 1000 bootstrap replications. The phylogenetic tree showed that three species of Aquilaria formed one monophyletic clade with 100% bootstrap value (). The complete chloroplast genome of A. crassna will provide a useful resource for the conservation genetics of this species as well as for the phylogenetic studies of Thymelaeaceae.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Barden A, Anak NA, Muliken T, Song M. 2000. Heart of the matter: agarwood use and trade and CITES Implementation for Aquilaria malaccensis. Cambridge, UK: TRAFFIC International.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Harvey-Brown Y. 2018. Aquilaria crassna. The IUCN Red List of Threatened Species 2018: e.T32814A2824513.
- Hashim YY, Kerr PG, Abbas P, Salleh HM. 2016. Aquilaria spp. (agarwood) as source of health beneficial compounds: A review of traditional use, phytochemistry and pharmacology. J Ethnopharmacol. 189:331–360.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Mudunuri SB, Nagarajaram HA. 2007. IMEx: imperfect microsatellite extractor. Bioinformatics. 23:1181–1187.
- Ng LT, Chang YS, Kadir AA. 1997. A review on agar (gaharu) producing Aquilaria species. J Trop Prod. 2:272–285.
- Patel RK, Jain M. 2012. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7:e30619.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 57:758–771.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
- Zhang YH, Huang Y, Li ZM, Zhang SD. 2019. Characterization of the complete chloroplast genome of the vulnerable agarwood tree, Aquilaria yunnanensis (Thymelaeaceae). Conserve Genet Resour. 11:161–164.