Abstract
The complete chloroplast genome of Sorghum bicolor subsp. drummondii cultivar Sa (a modern Sudan grass cultivar) was sequenced and analyzed in the present study. The chloroplast genome is 140,754 bp in length and includes a large single-copy region 82,688 bp in length, a small single-copy region 12,503 bp, and two inverted repeat regions 22,782 bp each. The genome contains 104 unique genes, including 4 rRNAs, 29 tRNAs, and 71 protein-coding genes. The phylogenetic analysis showed that Sudan grass cultivar Sa in a clade with five other complete chloroplast genomes of S. bicolor. The work facilitates studies on population genetic structure and phylogenetic relationships in genus Sorghum.
Sorghum bicolor subsp. drummondii, common name Sudan grass, is an important forage crop with remarkable drought tolerance (Creamer and Baldwin Citation2000). Sudan grass also has the potential to produce large amounts of biomass that builds soil quality in a short period of time. The plant also recycles nitrogen, outcompetes weeds, and reduces soil erosion (Acevedo et al. Citation2019). The cultivar Sa was widely planted in China because of its high biomass and excellent drought tolerance (Zhan et al. Citation2008). It is controversial about whether Sorghum and Sudan grass are the same species. Snowden (Citation1936) treated Sudan grass as S. sudanense, a species different from Sorghum in spikelet, anthotaxy and plant traits. De Wet and Huckabay (Citation1967) suggested that Sudan grass be placed as a subspecies, drummondii, of S. bicolor (L.) Moench. Zhan et al. (Citation2008) also suggested that Sudan grass should be placed as a Sorghum subspecies under S. bicolor based on SSR markers evidence. In this study, we characterized the complete chloroplast genome of Sorghum bicolor subsp. drummondii Sa and explored its phylogenetic relationship within the genus Sorghum. The results will facilitate future studies on population genetic structure and phylogenetic relationships.
The Sudan grass cultivar Sa (voucher APGRCFC-S00001) was harvested from the planting station of Anhui Science and Technology University, Fengyang county (32°52′ N, 177°33′ E), Anhui province, China. The voucher specimen and its DNA were deposited at the Anhui Provincial germplasm resource center for forage crops (http://www.ahstu.edu.cn/nxy/info/1021/7873.htm, Jieqin Li, [email protected]). Total DNA was isolated from fresh leaves using the DNAsecure Plant Kit (QiagenTM, Cat. No. DP320) following the manufacturer’s instructions. A library of circularized DNA fragments of 200–400 bp was amplified to make DNA nanoballs (DNBseqTM). Paired end 150 bp reads were generated on the MGISEQ-2000 sequencer. The raw data were filtered by SOAPnuke (Chen et al. Citation2018) with the filter parameters ‘-n 0.01 -l 20 -q 0.3 -A 0.25 –cutAdaptor -Q 2 -G –polyX 50 –minLen 150’. Then the complete chloroplast genome was assembled by GetOrganelle v1.7.4 (Jin et al. Citation2020). The genome sequences were annotated using GeSeq (Tillich et al. Citation2017) with the reference sequence of Sorghum. bicolor (NC_008602). The complete chloroplast genome sequence was submitted to NCBI under the accession number MW999225.
The complete chloroplast genome displayed the typical quadripartite structure found in most angiosperm chloroplast genomes (Luo et al. Citation2021), showing a high level of gene synteny to previously published Sorghum chloroplast genomes (Song et al. Citation2019). The genome includes a large single-copy (82,688 bp), a small single-copy (12,503 bp), and a pair of inverted repeats (22,782 bp). The GC content of chloroplast genome is 38%. The GC content for the large single-copy, the small single-copy, and the inverted repeats is 36%, 33%, and 44%, respectively. The genome encodes a total of 104 unique genes, of which there are 4 rRNAs s, 29 tRNAs and 71 protein-coding genes.
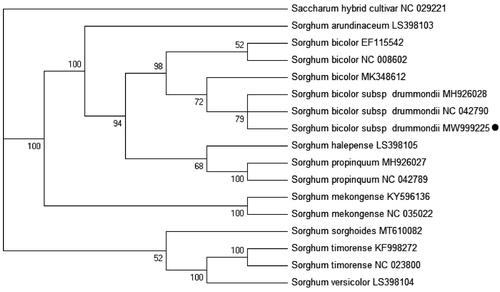
A phylogenetic analysis was performed based on complete chloroplast genome sequences from 16 Sorghum spp. and 1 Sachharum hybrid cultivar as the outgroup taxon. The 17 chloroplast genomes were aligned using MAFFT v7.313 with the auto settings (Katoh and Standley Citation2013). A Maximum likelihood phylogenetic tree was conducted using IQtree 1.68 (Nguyen et al. Citation2015) with the model K3Pu + F + I chosen according to the Bayesian analysis (BI) method and 1000 bootstrap replicates. Sudan grass cultivar Sa (S. bicolor subsp. drummondii) and S. bicolor were strongly resolved in the same clade (). These results also suggested that Sudan grass should be classified as a subspecies in S. bicolor as Zhan et al. (Citation2008) concluded using SSR markers.
Figure 1. Maximum-likelihood tree based the 17 complete chloroplast genome sequences. The number at each node are the bootstrap percentages. The number after the species name is the GenBank accession number. The dark circle represents the chloroplast genome of the Sorghum bicolor subsp.drummondii cultivar Sa analyzed here in this study.
Data availability
The genome sequence data that support the results of the study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the Accession no. MW999225. The associated BioProject and SRA are PRJNA718131 and SRR14089996, respectively. The Bio-sample number is APGRCFC-S00001in Anhui Provincial germplasm resource center for forage crops.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Acevedo A, Simister R, McQueen-Mason SJ, Gómez LD. 2019. Sudangrass, an alternative lignocellulosic feedstock for bioenergy in Argentina. PLOS One. 14(5):e0217435.
- Chen Y, Chen Y, Shi C, Huang Z, Zhang Y, Li S, Li Y, Ye J, Yu C, Li Z, et al. 2018. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience. 7(1):1–6.
- Creamer NG, Baldwin KR. 2000. An evaluation of summer cover crops for use in vegetable production systems in North Carolina. HortScience. 35(4):600–603.
- De Wet JMJ, Huckabay JP. 1967. The origin of Sorghum bicolor. II. Distribution and domestication. Evolution. 21(4):787–802.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Luo C, Huang WLue, Sun H, Yer H, Li X, Li Y, Yan B, Wang Q, Wen Y, Huang M, et al. 2021. Comparative chloroplast genome analysis of impatiens species (Balsaminaceae) in the Karst area of China: insights into genome evolution and phylogenomic implications. BMC Genomics. 22(1):571.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Snowden JD. 1936. The cultivated races of Sorghum. Tokyo: Adlard& Son, Ltd.; p. 274.
- Song Y, Chen Y, Lv J, Xu J, Zhu S, Li M. 2019. Comparative chloroplast genomes of Sorghum species: sequence divergence and phylogenetic relationships. Biomed Res Int. 2019:5046958.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Zhan QW, Zhang TZ, Wang BH, Li JQ. 2008. Diversity comparison and phylogenetic relationships of S. bicolor and S. sudanense as revealed by SSR markers. Plant Sci. 174(1):9–16.
