Abstract
Sinocyclocheilus anatirostris Lin and Luo, 1986 is a member of the endemic Chinese genus Sinocyclocheilus Fang, 1936, living in dark caves with absence of eyes and scales. Muscle tissue was collected from cavefish samples from Guangxi, China, and complete mitogenome was sequenced. This is the first report of the mitogenome of S. anatirostris. This mitogenome consists of 13 protein-coding genes (PCGs), two rRNA genes (12S rRNA and 16S rRNA), 22 tRNA genes, a control region (CR), and comprises 31.2% A, 24.4% T, 16.7% G, and 27.7% C bases. Phylogenetically, S. anatirostris is closely related to the Sinocyclocheilus furcodorsalis, and originated in the late Miocene, ∼6.07 Ma.
Introduction
The golden-line fish genus Sinocyclocheilus Fang Citation1936, endemic to the karst areas of southwest China, including Guangxi, Guizhou, Yunnan, and Hubei provinces, has 77 species recorded, 72 of which can be divided into five species groups (Fang Citation1936; Zhao and Zhang Citation2009; Wen et al. Citation2022; Xu et al. Citation2023), namely Sinocyclocheilus angularis, Sinocyclocheilus cyphotergous, Sinocyclocheilus microphthalmus, Sinocyclocheilus jii, and Sinocyclocheilus tingi groups. Among these five species groups, S. angularis and S. microphthalmus groups have evolved complex morphological characters adapted to the dark environment, such as horn-like structure on the back of the head (long, short, and forked), degeneration or absence of eyes and scales, and strengthening of locomotor and sensory organs (Ma et al. Citation2019). Currently, cavefishes of the S. angularis group are mainly distributed in the Nanpanjiang, Beipanjiang, and Hongshui rivers (), and the limited genetic information may limit our understanding of the evolutionary history.
Figure 1. Type locality of species of the S. angularis and S. microphthalmus groups within karst areas of Southwest China (a) and ecological photograph of S. anatirostris (by Jiajun Zhou) (b).
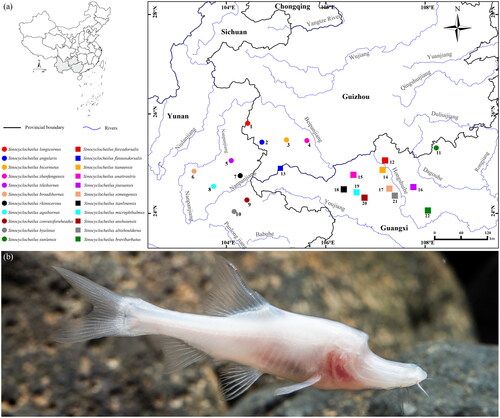
Sinocyclocheilus angularis Lin and Luo Citation1986 is a typical cave species within S. angularis group, with a duck-billed snout, absence of eyes, scales and body pigmentation, and a short horn-like structure on the back of the head () (Lin and Luo Citation1986; Zhao and Zhang Citation2009; Lan et al. Citation2013). This species has long been assessed as vulnerable by the International Union for Conservation of Nature (IUCN Citation2022) due to lack of sufficient recognition and protection. In this study, we sequenced the complete mitogenome of S. angularis and analyzed its phylogenetic relationships with other species of the congenus. The publication of this data is informative for the conservation of S. angularis and subsequent studies.
Materials
Single sample tissues of Sinocyclocheilus anatirostris were collected in 2018 from Youping Town, Leye County, Guangxi Zhuang Autonomous Region, China (24.99766151°N, 106.48204029°E; elevation: 575 m), and kept in Guizhou Normal University, Guiyang City, Guizhou Province, China (sample ID: GZNU-YZ01, http://gznu.edu.cn, contact person: Jiang Zhou; Email: [email protected]).
Methods
Genomic DNA for sample was extracted from 95% ethanol-preserved tissue using the cetyltrimethylammonium bromide method (Allen et al. Citation2006). Mitogenome was sequenced at TSINGKE Biotechnology Co., Ltd. (Chengdu, China) using an Illumina Novaseq 6000 platform (Illumina, USA) with 150 bp paired-end reads. Sequencing generated 4.9 G of raw data, which were filtered using SOAPnuke 1.3 (Chen et al. Citation2018) to obtain 4.9 G of clean data. Clean data were de novo assembled using Mitoz v. 2.3 software (Supplementary Material, Figure S1). The mitochondrial genome was spliced using SPAdes 3.13 (Bankevich et al. Citation2012; parameters: -k 127), and the spliced sequences were blast (version: BLAST 2.2.30+; parameters: -evalue 1e-5) with the reference mitochondrial genome to identify candidate sequences for assembly. The assembled mitogenome was annotated with genes using MITOS2 (Bernt et al. Citation2013) and uploaded to the National Center for Biotechnology Information (NCBI, USA) under the accession number OP429108. We downloaded 30 mitogenomes from NCBI for molecular analysis, including 28 species of Sinocyclocheilus and two outgroups. We used MAFFT 7.471 (Katoh and Standley Citation2013) implemented in PhyloSuite 1.2.2 (Zhang et al. Citation2020) for sequence alignment and PartitionFinder 2.1.1 (Lanfear et al. Citation2012) to select the best-fit model (GTR + I + R). In total, 2000 ultrafast bootstrap replicates were run in IQ-tree 2.0.4 to reconstruct the phylogenetic tree. Divergence times of the two node calibrations were analyzed using BEAST 2.4.7 (Bouckaert et al. Citation2014) as previously described (Wen et al. Citation2022).
Results
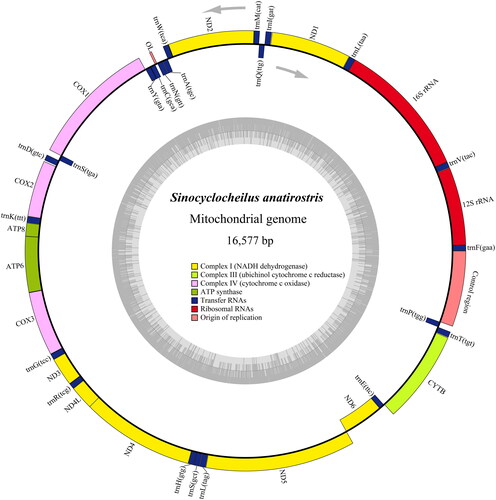
The complete mitogenome length of S. anatirostris was 16,577 bp (). The mitogenome contained a control region, 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, and two ribosomal RNA (rRNA) genes, which were 31.2% A, 24.4% T, 16.7% G, and 27.7% C. Among these genes, the ND6, trnQ(ttg), trnA(tgc), trnN(gtt), trnC(gca), trnY(gta), trnS(tga), trnE(ttc), and trnP(tgg) were encoded on the L-strand, while the remaining genes were encoded on the H-strand. Within the 13 PCGs, except for COI, which starts at a GTG codon, the codons of the remaining 12 PCGs started with ATG. The arrangement of these genes was similar to that of S. angularis (Luo and Zhang Citation2021). Except for ND2 and ATP8 with codon TAG, COX2, and ND4 with T– as stop codons, and COI with AGG as a stop codon, the remaining eight PCGs used TAA as a stop codon.
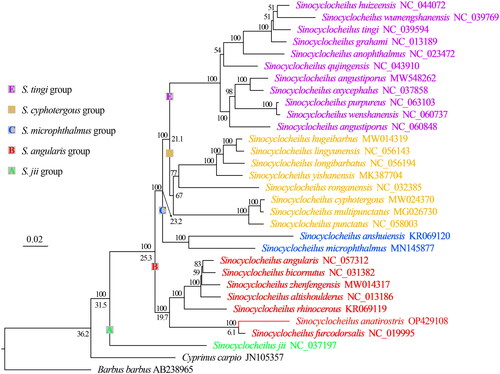
Phylogenetic tree reconstructed using the mitogenome shows that Sinocyclocheilus has six major clades corresponding to five species groups, but the monophyly of S. cyphotergous is not supported. Phylogenetic tree and divergence time suggests that S. anatirostris may be close to Sinocyclocheilus furcodorsalis and originated in the late Miocene, ∼6.07 Ma ().
Discussion and conclusions
The phylogenetic result support the division of Sinocyclocheilus into five species groups, i.e. S. jii, S. tingi, S. cyphotergous, S. angularis, and S. microphthalmus groups (Zhao and Zhang Citation2009; Wen et al. Citation2022; Xu et al. Citation2023), but S. cyphotergous group needs to be studied further using more species. Since the Miocene, the occurrence of orogeny and monsoonal climates in southwestern China have created favorable conditions for the speciation of the genus by geographic isolation (Yuan et al. Citation1995; Zhao and Zhang Citation2009; Wen et al. Citation2022), which has resulted in the distribution of different species of the genus in different river systems (). Sinocyclocheilus furcodorsalis and S. anatirostris have distinct horn-like structures on the back of their head, as well as body pigmentation absence (Lan et al. Citation2013), while S. angularis, Sinocyclocheilus bicornutus, Sinocyclocheilus altishoulderus, and Sinocyclocheilus zhenfengensis have long or short horn-like structures, but normal or slightly degraded body coloration (Zhao and Zhang Citation2009; Liu et al. Citation2018). Based on the time of divergence, we suggest that the loss of body coloration and the emergence of the most recent common ancestor of inconspicuous or short horns occurred 19.76 Ma ago, while further development of short horns into longer horns required at least 8 Ma. Thus, we hypothesize that at least one intensification event occurred during the evolution of the horns.
We present here the first report of the complete mitogenome of S. anatirostris, successfully sequenced, assembled, and annotated. Phylogenetic relationships and time of origin of this species based on the mitogenome level were assessed. The mitogenome published here can be used as a basis for further studies on the phylotaxonomy, conservation biology and biogeography of Sinocyclocheilus.
Ethical approval
The study was approved by the institutional review board of Guizhou Normal University, Guiyang, Guizhou, China. All samples were obtained following Chinese regulations for the implementation of the protection of terrestrial Wild Animals (State Council Decree (1992) No. 13), and the Guidelines for the Care and Use of Laboratory Animals by the Ethics Committee at Guizhou Normal university (Guiyang, Guizhou, China).
Vulnerable species statement
We have strictly followed the policies of the International Union for Conservation of Nature (IUCN) policies research involving species at risk of extinction, the Convention on Biological Diversity and the Convention on the Trade in Endangered Species of Wild Fauna and Flora.
Author contributions
Conceptualization: Xin-Rui Zhao, Huai-Qing Deng, and Jiang Zhou; specimen collection and identification: Xin-Rui Zhao, Chang-Ting Lan, Jing Yu, and Jia-Jun Zhou; data analysis: Xin-Rui Zhao, Chang-Ting Lan, Jing Yu, Xue-Li Lu, Ning Xiao, and Jiang Zhou; writing-original draft preparation: Xin-Rui Zhao, Xue-Li Lu, Huai-Qing Deng, Ning Xiao, and Jiang Zhou; writing-review: Xin-Rui Zhao, Huai-Qing Deng, and Jiang Zhou. All authors agree to be accountable for all aspects of the work.
Supplemental Material
Download TIFF Image (144.3 MB)Acknowledgments
We would like to thank Tao Luo for his assistance with software application and analysis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in the GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession No. OP429108. The associated BioProject, SRA, and Bio-Sample numbers are: PRJNA889191, SRR21856437, and SAMN31233307, respectively.
Additional information
Funding
References
- Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF. 2006. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc. 1(5):2320–2325.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Bouckaert R, Vaughan TG, Barido-Sottani J, Duchêne S, Fourment M, Gavryushkina A, Heled J, Jones G, Kühnert D, De Maio N, et al. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLOS Comput Biol. 10(4):e1003537.
- Chen Y, Chen Y, Shi C, Huang Z, Zhang Y, Li S, Li Y, Ye J, Yu C, Li Z, et al. 2018. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience. 7(1):1–6.
- Fang PW. 1936. Sinocyclocheilus tingi, a new genus and species of Chinese barbid fishes from Yunnan. Sinensia. 7:588–593.
- IUCN. 2022. The red list of threatened species. Version 2022-1. [accessed 2022 Aug 26]. http://www.iucnredlist.org.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Lan JH, Gan X, Wu TJ, Yang J. 2013. Cave fishes of Guangxi, China. Beijing: Science Press.
- Lanfear R, Calcott B, Ho SYW, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29(6):1695–1701.
- Lin RD, Luo ZF. 1986. A new blind barbid fish (Pisces, Cyprinidae) from subterranean water in Guangxi, China. Acta Hydrobio Sin. 10(4):380–382.
- Liu T, Deng HQ, Ma L, Xiao N, Zhou J. 2018. Sinocyclocheilus zhenfengensis, a new cyprinid species (Pisces: Teleostei) from Guizhou Province, Southwest China. J Appl Ichthyol. 34(4):945–953.
- Luo Q, Zhang RY. 2021. The complete mitochondrial genome and phylogenetic analysis of Sinocyclocheilus angularis (Cypriniformes: Cyprinidae). Mitochondrial DNA B Resour. 6(12):3438–3439.
- Ma L, Zhao YH, Yang JX. 2019. Cavefish of China. In: Encyclopedia of caves. Cambridge: Academic Press.
- Wen HM, Luo T, Wang YL, Wang SW, Liu T, Xiao N, Zhou J. 2022. Molecular phylogeny and historical biogeography of the cave fish genus Sinocyclocheilus (Cypriniformes: Cyprinidae) in southwest China. Integr Zool. 17(2):311–325.
- Xu C, Luo T, Zhou JJ, Wu L, Zhao XR, Yang HF, Xiao N, Zhou J. 2023. Sinocyclocheilus longicornus (Cypriniformes, Cyprinidae), a new species of microphthalmic hypogean fish from Guizhou, Southwest China. ZK. 1141:1–28.
- Yuan DX, Li B, Liu Z. 1995. Karst in China. ERA J. 18(1–2):62–65.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.
- Zhao YH, Zhang CG. 2009. Endemic fishes of Sinocyclocheilus (Cypriniformes: Cyprinidae) in China—species diversity, cave adaptation, systematics and zoogeography. Beijing: Science Press.