Abstract
The mitogenome of a soft coral, Eleutherobia rubra (Brundin, 1896), was completely sequenced for the first time. The total mitogenome length of E. rubra is 18,724 bp with 14 protein-coding genes, two ribosomal RNA genes, one transfer RNA gene (tRNA–Met), and one non-coding region (NCR). The gene order is also consistent with other Alcyoniidae species. The base composition is 30.1% A, 16.7% C, 19.5% G, and 33.7% T, with a G–C content of 36.2%. This is the first record of the complete mitogenome sequence of the genus Eleutherobia.
Introduction
Eleutherobia rubra (Brundin, 1896) belonging to Alcyoniidae is a finger-shaped azooxanthellate soft coral. Since Eleutherobia was first reported by Pütter in 1900, 13 species are known to date. Eleutherobia rubra inhabits the temperate waters around Korea, Japan, the USA (California), and Australia, at depths of 20–182 m (Song Citation1976; Verseveldt and Bayer Citation1988; Imahara et al. Citation2014). In Korea, this species occurs in a wide area around the western, southern, and eastern coasts, and forms several populations with high density, particularly in the coastal sea off the south coast of Korea (). However, these populations are threatened by global warming. The Kuroshio, which transports excess heat from tropical ocean to the north in the western North Pacific, has warmed twice to three times faster than the global average (Wang et al. Citation2016; Lam et al. Citation2021; Sasaki and Umeda Citation2021; Wan et al. Citation2023). Consequently, the East Asian marginal seas including Korean peninsula, which are strongly affected by the Kuroshio Current, have become rapidly warming regions (Wang and Wu Citation2022; Lee et al. Citation2023). Recently, loss of these populations and changes in transcription and symbiotic bacterial composition have been observed due to thermal stress (Lee et al. Citation2020, Citation2023). Coral bleaching and mass mortality due to heatwaves have also been observed worldwide, including in the Mediterranean and the Great Barrier Reefs (Hughes et al. Citation2017; Garrabou et al. Citation2022; McGowan and Theobald Citation2023).
Figure 1. Eleutherobia rubra. (A) Population in the coastal sea off the south coast of Korea. (B) Voucher specimen used in this study. Photographs of habitat and voucher were taken by Seung-Hwan Park (underwater photographer at H Dive) and Chi-Hyeon Kim, respectively, and copyright license agreements were obtained.
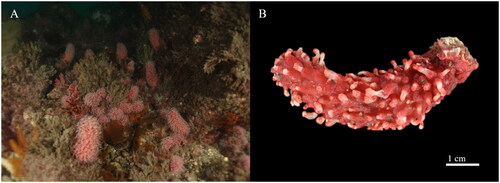
Populations with rapidly decreasing population sizes are easily at risk of extinction due to loss of genetic diversity (Kliman et al. Citation2008). Genetic management of species at risk of potentially endangered and resolving taxonomic uncertainties using genetic markers are important factors in conservation genetics (Frankham Citation2019; Hoban et al. Citation2023). Therefore, as the beginning of efforts to conserve the E. rubra population by identifying and managing genetic diversity using mitochondrial markers, the E. rubra mitogenome was analyzed. Approximately, 3,290 species of octocorals are known worldwide, but mitogenome data have been reported from only 6.7% (about 221 species) species so far (NCBI Citation2023; WoRMS Editorial Board Citation2023). This is the first report of the complete mitogenome in Eleutherobia. In addition, these data will also provide valuable information for further studies on the molecular taxonomy and phylogeny of octocorals, which are problematic in their taxonomy due to limited range of morphological characters, and insufficient and inadequate descriptions (Kessel et al. Citation2023).
Materials and methods
One specimen of E. rubra was collected from the subtidal zone of Eoyu Island in the coastal sea off the south coast of Korea (34°39′34.58″ N, 128°34′31.43″ E), and deposited at the Cnidaria Bioresources Bank of Korea, Woosuk University, Jincheon, South Korea (voucher number: CBB17CnAnE226, Prof. Sung-Jin Hwang, [email protected]). For identification, detailed morphological characteristics of the sclerites were confirmed following Song (Citation1976).
Total genomic DNA was extracted from the polyp tissue of the voucher specimen using the phenol–chloroform method using a lysis buffer containing high concentration of urea (10 mM Tris–HCl, pH 8.0; 125 mM NaCl; 10 mM EDTA, pH 8.0; 1% SDS; 8 M urea) developed by Asahida et al. (Citation1996). The complete mitogenome sequence was amplified using long-range PCR (LR-PCR), and then three LR-PCR products covering whole mitochondrial genome were sequenced by the primer walking method with 26 primers newly designed in this study (Table S1 for primers and Figure S1 for PCR gel image). The PCR reaction solutions were made using AccuPower ProFi Taq PCR PreMix (Bioneer, Daejeon, South Korea), and PCR amplification was performed according to the user manual of the ProFlex PCR System (Life Technologies, Carlsbad, CA). LR-PCR products were directly sequenced using 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA).
The complete circular mitogenome sequence of E. rubra was assembled starting with COX1 partial sequences as the first location in the gene map, the circular form was determined by identifying overlapped sequences between the first and the last partial sequences and compared the completed sequence with other mitogenome sequences in Alcyoniidae species. All LR-PCR sequences were checked and corrected during the assembly process using Geneious 9.1.8 (Biomatters, Auckland, New Zealand) (Kearse et al. Citation2012). The 14 protein-coding genes (PCGs) were annotated by identifying their open reading frames (ORFs), and by comparing them with other reported mitogenomes of Alcyoniidae species using MITOS web server (Bernt et al. Citation2013). The two ribosomal RNA genes (rRNAs) and one transfer RNA gene were determined by comparison with homologous gene sequences of other Alcyoniidae mitochondrial genomes.
The phylogenetic analysis was performed using mitogenomes sequences of eight previously published Alcyoniidae (Muthye et al. Citation2022) and the E. rubra in this study. Two Xeniidae mitogenomes were used as outgroup. Mitogenome sequences (14 PCGs nucleotides, excluding stop codons) of a total 11 species including E. rubra were concatenated and aligned using the multiple sequence alignment program, MAFFT v.7 (Katoh and Standley Citation2013) for phylogenetic analysis. A phylogenetic tree was reconstructed based on the concatenated dataset using the maximum-likelihood (ML) method with the GTR + G + I model in raxmlGUI 2.0 (Edler et al. Citation2021), with the bootstrap values being calculated from 10,000 replicates.
Results
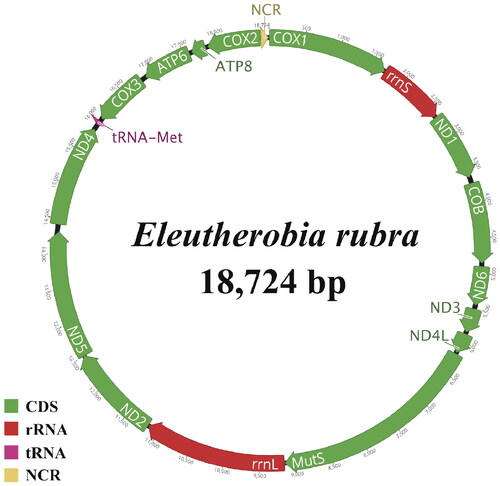
The complete circular mitogenome of E. rubra was 18,724 bp in length (GenBank accession no. ON814482) (), which was within published mitogenome lengths for Alcyoniids (18–19 kb). MutS gene, which is involved in DNA repair in octocoral mitochondria, was found in E. rubra mitogenome. Recently, the gene was utilized as a highly effective molecular marker for phylogenetic analysis in the octocoral group (McFadden et al. Citation2022; Muthye et al. Citation2022). E. rubra mitogenome has 17 genes (14 PCGs, two rRNAs, and one transfer RNA gene (tRNA–Met)) and one non-coding region (NCR) of 109 bp. The gene order shows an ancestral octocoral mt gene order, consistent with other Alcyoniidae species (Figueroa and Baco Citation2015). Regarding its nucleotide base composition, this mitogenome has A, C, G, and T contents of 30.1%, 16.7%, 19.5%, and 33.7%, respectively, with a G + C content of 36.2%. Two rRNAs (rrnS and rrnL) and 10 PCGs were encoded by the heavy strand. Four PCGs (ATP6, ATP8, COX2, and COX3) and tRNA–Met were encoded by the light strand. All PCGs began with ATG as a start codon. In addition, all PCGs used TAR as a stop codon. Ten genes (ATP8, COX1–3, COB, ND1–3, ND5, and ND6) ending with TAG and four genes (ATP6, MutS, ND4, and ND4L) ended with TAA. The two rRNA genes contained rrnS and rrnL between COX1 and ND1 or between MutS and ND2. NCR was located between COX1 and COX2.
Figure 2. Circular representation of the complete mitogenome for E. rubra. The genes were colored based on their functional groups. Arrows show the directions of transcription.
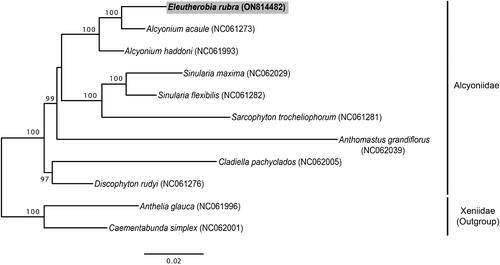
Phylogeny analysis indicated that this new mitogenome sequence of genus Eleutherobia clustered with Alcyonium species in family Alcyoniidae (). The tree showed that E. rubra and A. acaule had a close relationship with a very high nodal support (100% BP in ML). E. rubra and A. acaule shared sequence similarities of 98% for the total length of mitogenome sequences (18,724 bp of A. acaule). According to a previous study (McFadden et al. 2022), Eleutherobia species and A. acaule belonged to a sister group in phylogenetic analysis using MutS gene, similar to results of this study.
Figure 3. Maximum-likelihood tree based on concatenated 14 PCGs sequence dataset from 11 Malacalcyonacea species. Two Xeniidae species (Anthelia glauca and Caementabunda simplex) were used as outgroups. GenBank accession numbers of each sequence are marked behind their corresponding species names in the tree. Sequence obtained in this study is in bold.
Discussion and conclusions
Through this study, we report the first complete mitogenome sequencing with E. rubra for the genus Eleutherobia. The results of this study can be used for the population conservation of E. rubra by identifying and managing genetic diversity using mitochondrial genetic markers, and for comprehensive taxonomic studies of the Alcyoniidae including this species as a representative of the genus Eleutherobia.
Author contributions
Chi-Hyeon Kim: data analysis and manuscript writing. Sang-Hwa Lee: mapping and phylogenetic analysis. In-Young Cho: identification and manuscript revising. Min-Seop Kim: manuscript reviewing. Seonock Woo: manuscript reviewing. Keun-Yong Kim: data collection and analysis. Sung-Jin Hwang: designing this study, manuscript reviewing, and approval of final version to be published.
Ethical approval
This research does not involve ethical research. However, the sample collection area was designated and protected as Hallyeohaesang National Park, so the sample was collected with permission from the Korea National Park Service. E. rubra is widespread in South Korea, and is not listed as a threatened or endangered species.
Supplemental Material
Download JPEG Image (11.7 MB)Supplemental Material
Download MS Word (21.5 KB)Disclosure statement
The authors declare no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The data that support the findings of this study are openly available in the GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession ON814482. The associated BioProject, SRA and Bio-sample numbers are PRJNA930047, SRR24682002, and SAMN29133432, respectively.
Additional information
Funding
References
- Asahida T, Kobayashi T, Saitoh K, Nakayama I. 1996. Tissue preservation and total DNA extraction from fish stored at ambient temperature using buffers containing high concentration of urea. Fish Sci. 62(5):727–730. doi: 10.2331/fishsci.62.727.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi: 10.1016/j.ympev.2012.08.023.
- Edler D, Klein J, Antonelli A, Silvestro D, Matschiner M. 2021. raxmlGUI 2.0: a graphical interface and toolkit for phylogenetic analyses using RAxML. Methods Ecol Evol. 12(2):373–377. doi: 10.1111/2041-210X.13512.
- Figueroa DF, Baco AR. 2015. Octocoral mitochondrial genomes provide insights into the phylogenetic history of gene order rearrangements, order reversals, and cnidarian phylogenetics. Genome Biol Evol. 7(1):391–409. doi: 10.1093/gbe/evu286.
- Frankham R. 2019. Conservation genetics. In: Fath B, editor. Encyclopedia of ecology. 2nd ed., Vol. 1. Amsterdam: Elsevier; p. 382–390.
- Garrabou J, Gómez‐Gras D, Medrano A, Cerrano C, Ponti M, Schlegel R, Bensoussan N, Turicchia E, Sini M, Gerovasileiou V, et al. 2022. Marine heatwaves drive recurrent mass mortalities in the Mediterranean Sea. Glob Change Biol. 28(19):5708–5725. doi: 10.1111/gcb.16301.
- Hoban S, Bruford MW, da Silva JM, Funk WC, Frankham R, Gill MJ, Grueber CE, Heuertz M, Hunter ME, Kershaw F, et al. 2023. Genetic diversity goals and targets have improved, but remain insufficient for clear implementation of the post-2020 global biodiversity framework. Conserv Genet. 24(2):181–191. doi: 10.1007/s10592-022-01492-0.
- Hughes TP, Kerry JT, Álvarez-Noriega M, Álvarez-Romero JG, Anderson KD, Baird AH, Babcock RC, Beger M, Bellwood DR, Berkelmans R, et al. 2017. Global warming and recurrent mass bleaching of corals. Nature. 543(7645):373–377. doi: 10.1038/nature21707.
- Imahara Y, Iwase F, Namikawa H. 2014. The octocorals of Sagami bay. Tokyo, Japan: Tokai University Press; p. 1–398.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. doi: 10.1093/bioinformatics/bts199.
- Kessel GM, Alderslade P, Bilewitch JP, Schnabel KE, Gardner JP. 2023. The use of integrative taxonomy in Octocorallia (Cnidaria: Anthozoa): a literature survey. Zool J Linn Soc. 198(2):677–690. doi: 10.1093/zoolinnean/zlac099.
- Kliman R, Sheehy B, Schultz J. 2008. Genetic drift and effective population size. Nat Educ. 1(3):3.
- Lam AR, MacLeod KG, Schilling SH, Leckie RM, Fraass AJ, Patterson MO, Venti NL. 2021. Pliocene to earliest Pleistocene (5–2.5 Ma) reconstruction of the Kuroshio Current Extension reveals a dynamic current. Paleoceanogr Paleoclimatol. 36(9):e2021PA004318.
- Lee NY, Yum SS, Woo SO. 2020. Differentially expressed genes of octocoal, Eleutherobia rubra against heat stress and the local environment. PICES-2020 Virtual Annual Meeting [online]. PICES. https://sciwatch.kiost.ac.kr/handle/2020.kiost/37662.
- Lee NY, Jo YJ, Woo SO. 2023. Symbiotic communication of octocoral, Eleutherobia rubra responding to environmental stress. EMBL Symposium. EMBL; p. 75. https://sciwatch.kiost.ac.kr/handle/2020.kiost/43981.
- Lee S, Park MS, Kwon M, Park YG, Kim YH, Choi N. 2023. Rapidly changing East Asian marine heatwaves under a warming climate. JGR Oceans. 128(6):e2023JC019761. doi: 10.1029/2023JC019761.
- McFadden CS, van Ofwegen LP, Quattrini AM. 2022. Revisionary systematics of Octocorallia (Cnidaria: Anthozoa) guided by phylogenomics. Bull Soc Syst Biol. 1(3):е8735.
- McGowan H, Theobald A. 2023. Atypical weather patterns cause coral bleaching on the Great Barrier Reef, Australia during the 2021–2022 La Niña. Sci Rep. 13(1):6397. doi: 10.1038/s41598-023-33613-1.
- Muthye V, Mackereth CD, Stewart JB, Lavrov DV. 2022. Large dataset of octocoral mitochondrial genomes provides new insights into mt-mutS evolution and function. DNA Repair. 110:103273. doi: 10.1016/j.dnarep.2022.103273.
- [NCBI] National Center for Biotechnology Information [Internet]. 2023. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988] – [accessed Jul 25]. https://www.ncbi.nlm.nih.gov/.
- Sasaki YN, Umeda C. 2021. Rapid warming of sea surface temperature along the Kuroshio and the China coast in the East China Sea during the twentieth century. J Clim. 34(12):4803–4815. doi: 10.1175/JCLI-D-20-0421.1.
- Song JI. 1976. A study on the classification of the Korean Anthozoa: 2. Alcyonacea. Korean J Syst Zool. 19(2):51–62.
- Verseveldt J, Bayer FM. 1988. Revision of the Genera Bellonella, Eleutherobia, Nidalia and Nidaliopsis (Octocorallia: Alcyoniidae and Nidalliidae), with descriptions of two new Genera. Zool Verh. 245(1):1–131.
- Wang YL, Wu CR, Chao SY. 2016. Warming and weakening trends of the Kuroshio during 1993–2013. Geophys Res Lett. 43(17):9200–9207. doi: 10.1002/2016GL069432.
- Wang YL, Wu CR. 2022. Rapid surface warming of the Pacific Asian marginal seas since the late 1990s. JGR Oceans. 127(12):e2022JC018744. doi: 10.1029/2022JC018744.
- WoRMS Editorial Board. 2023. World register of marine species. VLIZ; [accessed 2023 Jul 27]. https://www.marinespecies.org.
- Wan S, Xiang R, Steinke S, Du Y, Yang Y, Wang S, Wang H. 2023. Impact of the Western Pacific Warm pool and Kuroshio dynamics in the Okinawa Trough during the Holocene. Glob Planet Change. 224:104116. doi: 10.1016/j.gloplacha.2023.104116.
