Abstract
Rhamnus leptacantha C.K.Schneid. (1914). is a rare shrub species of the genus Rhamnus. The complete plastid genome of Rhamnus leptacantha was sequenced for the first time in this study. The total length of this genome is 161,248 bp with a large single copy (LSC) region (89,386 bp), a small single copy (SSC) region (19,000 bp), and two inverted repeat regions (IRs, 26,431 bp). A total of 133 functional genes were annotated, including 88 protein-coding genes, 37 tRNA genes and 8 rRNA genes. Plastome of R. leptacantha displayed a conservative structure and gene order. Phylogenetic analysis strongly supported R. leptacantha clustered with other members of genus Rhamnus. This study provides a foundation for further investigation of the complete chloroplast genome in inferring the evolution within the Rhamnaceae family.
Introduction
Rhamnus leptacantha, belonging to the genus Rhamnus of the Rhamnaceae family, is a rare and unique shrub species in China. It grows primarily in the temperate biome and is found only in forests and hillside shrublands in south-central and southwestern China. It has an upright form with small, dark green leaves and clusters of small, yellow-green flowers (Chen and Zhou Citation1982). Rhamnus leptacantha has been used for its medicinal properties, including as an anti-inflammatory, antispasmodic, and diuretic. As for the species of genus Rhamnus, the fruit of R. leptacantha can be used to make dyes, providing Chinese green, a dye used to impart a bright green color to silk and wool (Brunello Citation1973). However, studies on the chloroplast genome of genus Rhamnus even in Rhamnaceae family are still lacking. In this study, we fist report the complete plastid genome or chloroplast(cp) genome of R. leptacantha. Knowledge on the genome of R. leptacantha will be useful for studying the conservation of this species and for recovering its phylogenetic position within the genus Rhamnus and Rhamnaceae family.
Materials and methods
Plant material, DNA extraction and sequencing
Fresh and healthy leaf tissues of R. leptacantha were collected from Wuyuan Cave Scenic Spot, Badong County, Enshi City, Hubei Province, China (; 110°25′18.60′′E, 31°01′536.00′′N). The voucher specimens (No. 210503011) were deposited in the herbarium of Yangtze River Biodiversity Research Center (Jinhua Wu, [email protected]). The whole genomic DNA was isolated with a modified CTAB method (Doyle and Doyle Citation1987). Paired-end (PE) sequencing was conducted on the Illumina HiSeq X-Ten instrument at Beijing Genomics Institute (BGI) in Wuhan, China.
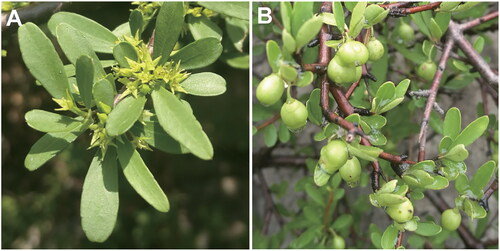
Figure 1. The characteristic images of R. leptacantha (shrubs spreading, dioecious; leaves alternate, small, narrow; flowers yellow-green, unisexual, 4-merous; drupes brown, subglobose. In this study, these pictures were taken by the author from Guiyun Huang). (A) Flowers, (B) Fruits. The voucher specimen (No. 210503011; 110°25′18.60′′E, 31°01′536.00′′N) was deposited in the herbarium of Yangtze River Biodiversity Research Center. (Jinhua Wu, [email protected]).
Genome assembly and annotation
We employed GetOrganelle pipeline (Jin et al. Citation2020) to assemble the plastome, and followed the parameter with -R 15 -k 65,85,105, then we used BWA-MEM algorithm of bwa v.0.7.17 (Li Citation2013) to compare the Illumina short sequence with the chloroplast genome sequence and SAMtools v1.7 (Li Citation2013) to calculate the coverage depth. The chloroplast genome was annotated by using PGA program (Qu et al. Citation2019) and CPGAVAS2 (Shi et al. Citation2019) with default parameters. CPGView (Liu et al. Citation2023) (http://www.1kmpg.cn/cpgview/) was used to check the accuracy of these genes. We also employed Geneious v.9.1.8 (Kearse et al. Citation2012) to verify the accuracy of the assembly, and to correct the start/stop codons and intron/exon boundaries of the annotation. The annotated plastome was drawn by the online tool CPGview with default parameters (http://47.96.249.172:16085/cpgview/drawmap) (). Finally, the annotated result was deposited in GenBank (accession number: OR346919).
Phylogenetic analysis
To reconstruct the phylogenetic position of R. leptacantha, we combined our newly generated data with seven previously published plastid genomes which representing the outgroup of genus Ventilago and other members of genera Rhamnus and Frangula (). We aligned 76 protein-coding genes using MAFFT V.7.308 (Katoh and Standley Citation2013) with default parameters and manually adjusted misaligned regions using Geneious. We reconstructed the maximum likelihood phylogeny based on a concatenation of the 76 genes using IQ-TREE (Nguyen et al. Citation2015) with the TVM + F + I substitution model as suggested by ModelFinder (Kalyaanamoorthy et al. Citation2017).
IR boundary analysis and rearrangement analysis
To test the length and variation of LSC/SSC regions in the chloroplast genome of genus Rhamnus, we employed IRscope (Amiryousefi et al. Citation2018) to generate the boundaries of the IR, SSC, and LSC regions. Furthermore, we applied Mauve 2.3.1 (Darling et al. Citation2004) to compare the plastome structures and gene rearrangements.
Results
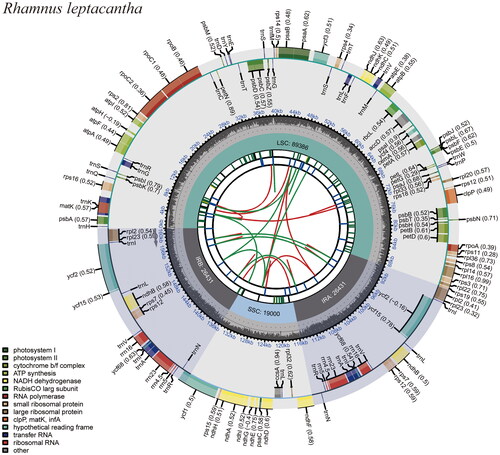
After examining the genome sequence for completeness, coverage depth and collinearity, the results showed that the plastome was reliable (Figure S1). Analysis of the genome annotation results also showed that the cis- and trans-splicing gene annotations were correct (Figure S2). The circular complete plastid genome of R. leptacantha was 161,248 bp in length with a typical quadripartite organization: a large single copy (LSC) region of 89,386 bp, a small single copy (SSC) region of 19,000 bp, respectively, and these two regions were separated by two inverted repeat regions (IRA and IRB), each 26,431 bp in length. A total of 133 functional genes were recovered, comprising 88 protein-coding genes, 37 tRNA genes and 8 rRNA genes. The overall GC content of the whole plastome is 37.1% ().
Figure 2. The genome map of R. leptacantha. Circular representation of the R. leptacantha chloroplast genome, showing the clockwise (genes inside the circle) and counterclockwise (outside) transcribed genes. Colors identify genes from the same functional category, following the figure legends. In the inner circle, the dark and light grey bars indicate the guanine + cytosine and adenine + thymine content, respectively. IRa and IRb: inverted repeat regions; LSC: large single copy region; SSC: small single copy.
The chloroplast genomes are relatively conserved in angiosperms in terms of size, structure and gene content, ranging from 120 kb to 170 kb in length in most cases (Shaw et al. Citation2007). The comparison of plastome structure and gene rearrangement suggested that the genes and their order in the chloroplast genome of R. leptacantha are the same as the Rhamnaceae family (Figure S3), which indicated that the R. leptacantha has a conserved chloroplast genome of structure. In addition, the result of IR boundary analysis also showed that the plastid genome of R. leptacantha presents a conserved length and IR boundaries (). In the chloroplast genome of R. leptacantha, ycf1 was located within the SSC/IR boundary, which is similar to other species in Rhamnaceae family except Frangula crenata. And all the plastome showed that the rpl2 and trnN were located outside the SSC/IR boundary. We also observed that the ndhF was located within the SSC/IR boundary in the plastomes of R. globosa and R. taquetii, while other plastomes were not.
Figure 3. (A) Phylogenetic tree reconstructed by maximum likelihood (ML) method based on 76 protein-coding genes from the eight Rhamnaceae chloroplast genomes. Numbers above the lines represent ML bootstrap values. (B) Comparative analyses of the boundary regions (LSC, SSC, and IR) and adjacent genes among eight chloroplast (cp) genomes. Gene names are shown in boxes, and their lengths in junction sites are displayed below the boxes, plus signs indicate genes located within the boundary, arrows indicate genes located outside the boundary. The following sequences were used: Rhamnus globosa_NC057506, Rhamnus heterophylla_NC057481, Rhamnus taquetii_NC045855, Frangula alnus_NC068506, Rhamnus crenata_LC635131, Ventilago harmandiana_NC065258, Ventilago leiocarpa_NC053785, 7 previously published species combined with our newly generated species Rhamnus leptacantha for a total of 8 species.

The phylogenetic analysis strongly suggested that R. leptacantha is sister to the rest of Rhamnus, and our phylogenetic analysis also recovered the differentiation of genus Rhamnus and genus Frangula, with a strong bootstrap support provide the sister relationship between genus Rhamnus and Frangula (), which supported the division of these two genera (Yu et al. Citation2008).
Conclusion and discussion
In this study, we fist report the complete chloroplast genome of R. leptacantha, and characterize its genomic features, which displayed a typical quadripartite structure (), with a conservative plastome structure and gene order (Figure S3). Phylogenetic analysis also determined its systematic position with other species of genus Rhamnus, and our phylogenetic results also supported the division of genus Rhamnus and Frangula (Yu et al. Citation2008).
Rhamnaceae, which includes 52 genera and more than 900 species of shrubs and trees, is distributed worldwide. Due to the lack of genome sequences, phylogenetic studies of Rhamnaceae family have been limited. Our findings suggest the potential benefit of complete plastid genomes in inferring the phylogenetic relationships within the Rhamnaceae family. Developing some bar-coding markers or SSRs based on our plastid genome can also be useful and applied in population genetics and informative in detecting genetic polymorphisms at population level. And the genetic resource information generated in this study will be valuable for further investigations into the biology and conservation breeding of Rhamnus. And we would expect a better resolved and more strongly supported phylogeny of Rhamnaceae in the near future when a more comprehensive sampling is available.
Author contributions
GY, LL, XW, DW, JW directed the study and designed the experiments. GY, LY and YW performed the data processing. GY, DW, and JW drafted the manuscript. All authors have revised and approved the final draft.
Ethical approval
This study includes no human, animal, or endangered plant samples. And the sample used in this study, including the collection of plant material has been carried out in accordance with guidelines provided by our institution. Field studies in our manuscript have complied with local legislation and appropriate permissions/license were granted while taking samples from a preserved/protected land.
Supplemental Material
Download TIFF Image (2.3 MB)Supplemental Material
Download TIFF Image (3.5 MB)Supplemental Material
Download TIFF Image (4.6 MB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. OR346919. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA998250, SRR25412703, and SAMN36701180, respectively.
Additional information
Funding
References
- Amiryousefi A, Hyvönen J, Poczai P. 2018. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 34(17):3030–3031. doi:10.1093/bioinformatics/bty220.
- Brunello F. 1973. The art of dyeing in the history of mankind. Vincenza, Italy: Neri Pozza Editore; p. 381.
- Chen YL, Zhou BK. 1982. Rhamnus. In: Chen YL, editor. Flora of China. Vol. 48(1). Beijing & Missouri Botanical Garden, St. Louis: Science Press; p. 019.
- Darling ACE, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14(7):1394–1403. doi:10.1101/gr.2289704.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi:10.1186/s13059-020-02154-5.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, Haeseler AV, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi:10.1038/nmeth.4285.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. doi:10.1093/bioinformatics/bts199.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079. doi:10.1093/bioinformatics/btp352.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi:10.1111/1755-0998.13729.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997. Available from: http://arxiv.org/abs/1303.3997.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi:10.1093/molbev/msu300.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Meth. 15(1):50. doi:10.1186/s13007-019-0435-7.
- Shaw J, Lickey EB, Schilling EE, Small RL. 2007. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Am J Bot. 94(3):275–288. doi:10.3732/ajb.94.3.275.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):65–73.
- Yu H, Ye HG, Xia NH. 2008. Taxonomic treatment of Chinese Frangula Mill. (Rhamnaceae). J Trop Subtrop Bot. 16(4):366–369.
