Abstract
The Ras pathway is a major driver in lung adenocarcinoma: over 75% of all cases harbor mutations that activate this pathway. While spectacular clinical successes have been achieved by targeting activated receptor tyrosine kinases in this pathway, little, if any, significant progress has been achieved targeting Ras proteins themselves or cancers driven by oncogenic Ras mutants. New approaches to drug discovery, new insights into Ras function, new ways of attacking undruggable proteins through RNA interference and new ways of harnessing the immune system could change this landscape in the relatively near future.
1. Lung adenocarcinoma is usually driven by the Ras pathway
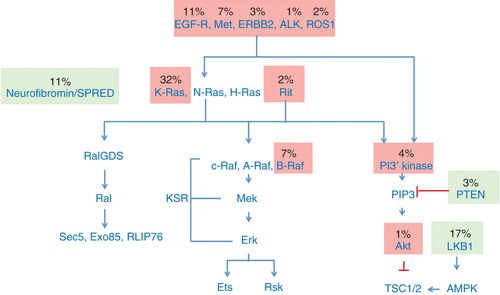
Ras proteins play a major role in lung adenocarcinoma, as judged by their frequent rate of mutation. K-Ras is activated by mutation in 32% of cases, and wild-type Ras proteins are activated by the loss of their negative regulator, neurofibromin, in another 11%. In addition, EGF-receptor (11%) and other tyrosine kinases (together, about another 10%) are activated by mutation or by over-expression, and signal through Ras proteins, as well as through other pathways. Taken together, at least 76% of all lung adenocarcinomas appear to be driven by aberrant signaling through Ras proteins. This number will likely increase as more drivers are detected: for example, the discovery that 2% adenocarcinomas of the lung may be driven by the Ras-related protein Rit1, underscores the fact that the contribution of Ras proteins may still not be fully appreciated () Citation[1].
2. The clinical need
Targeting receptor tyrosine kinases (RTKs) directly has proven a successful strategy, with remarkable, unprecedented clinical responses that result in dramatic improvements in quality and duration of life relative to previous protocols. A better understanding of mechanisms of drug resistance is likely to extend these major improvements substantially further Citation[2]. However, patients suffering from K-Ras-driven cancers do not benefit from these therapies, and, indeed, may progress faster during treatment with RTK inhibitors. A similar situation has been reported for patients suffering from malignant melanoma: patients whose tumors are driven by N-Ras mutations do not benefit from Raf kinase inhibitors such as vemurafenib and may indeed progress faster on therapy Citation[3]. Failure of Ras mutant cancers to respond to RTK therapy is not at all surprising, as RTKs are upstream of Ras: Ras mutant cancers are therefore less dependent on RTK signaling than normal cells or cells driven by mutant RTKs. Furthermore, persistent signaling from mutant Ras proteins leads to feedback inhibition of upstream signals, further contributing to loss of dependence on these targets. However, Ras cancers still retain some dependency on these signals Citation[4]; the strength of these signals is diminished, and therefore not a suitable point of therapeutic intervention. It is therefore abundantly clear that new strategies need to be developed to target K-Ras driven cancers, from which about one million people die every year.
3. Ways of targeting Ras directly
Therapeutic approaches to Ras cancers could involve the development of compounds that bind to Ras proteins directly, such as compounds described by Ostrem et al., described below Citation[5]. They could affect Ras processing or other post-translational modifications, or they could target downstream effectors of Ras, such as proteins in the Raf MAPK pathway. Less direct approaches might include exploitation of metabolic changes in Ras-driven cells, epigenetic changes associated with Ras activity, attack by harnessing the immune system or attack by delivery of siRNA that knock-out K-Ras expression or by taking advantage of viruses that replicate selectively in cancers in which the Ras pathway is active. All of these approaches are currently being tested, mostly in pre-clinical settings Citation[3]. The focus of this perspective will be the direct attack on Ras proteins themselves and the delivery of siRNA that knocks down Ras expression.
H-Ras, N-Ras and K-Ras proteins play essential roles in normal cells, many of which are redundant. Therefore, it is likely that ablation of all Ras functions by drugs that block Ras proteins indiscriminately would not be tolerated, though this has not been proven formally. Ideally, drugs would be developed that block oncogenic variants specifically. Alternatively, drugs that block K-Ras, and not H-Ras and N-Ras are likely be to effective, even though they do not distinguish between oncogenic K-Ras and wild-type K-Ras. This is because K-Ras, H-Ras and N-Ras play redundant roles in normal tissues, so that ablation of any one sub-type is expected to be tolerable.
The G12C mutant is the most common K-Ras mutant in lung adenocarcinoma, because the G > T transversion that gives rise to this amino acid change is a signature of exposure to tobacco smoke. The frequency of this mutation is higher in women than in men, particularly younger women with a lesser smoking history, for reasons which are not clear. Interestingly, activating mutations in EGF-receptor in lung carcinoma are also more common in women, but these cases are generally in never smokers Citation[6].
To target the K-Ras G12C protein directly, Ostrem and colleagues in Dr Kevan Shokat’s laboratory exploited the unique chemical properties of the cysteine at codon 12: this amino acid is uniquely nucleophilic, making it vulnerable to electrophilic attack. Using this insight, they developed small molecules that bind covalently to the cysteine-12 residue, and inactivate K-Ras as a result. These compounds only bind to the inactive, guanosine diphosphate (GDP) bound form of K-Ras. At first sight, this might appear to be the wrong target, as the active form of K-Ras is its guanosine triphosphate (GTP) bound state. Furthermore, activating mutations in K-Ras, such as those at codon 12, tend to lock K-Ras in this active state through inhibition of GTPase activating protein (GAP) mediated GTP hydrolysis. However, oncogenic mutants of Ras do hydrolyze GTP to GDP through their intrinsic GTPase mechanism. This is an extremely slow reaction. The GDP-bound form then returns slowly to the GTP-bound state when GDP dissociates and GTP binds. This reaction is also very slow, and indeed, levels of K-Ras in the GTP and GDP states are comparable for many oncogenic mutants. By binding covalently to the GDP-bound form of K-Ras G12C, conversion back to the GTP bound state is impossible, and eventually all the mutant protein becomes inactive.
The other common mutations in K-Ras (G12D, G12V, G13D) do not present equivalent opportunities for chemical attack. However, one encouraging outcome from the Shokat study, and similar efforts from others, is the appreciation that the surface of Ras proteins may have pockets in which a small molecule could bind, that had been missed in earlier X-Ray structures. Furthermore, each of these mutations perturbs the structure of the Ras protein in different ways, and a thorough analysis of each of mutant by crystallographic techniques as well as nuclear magnetic resonance and other biophysical approaches may reveal new opportunities. Surprisingly, no structures of oncogenic Ras proteins complexed with effectors (such as Raf kinase) or regulators (such as GAPs) are available. A thorough analysis of these structures is underway at the Frederick National Labs in the hope that new ways of targeting these proteins directly will emerge Citation[3]. All structures solved to date represent fragments of Ras proteins or unprocessed proteins. The precise way in which Ras proteins interact with the plasma membrane is unclear: this is important for several reasons: direct interaction of Ras with the membrane lipids may promote allosteric changes that alter how Ras proteins interact with their effectors. Furthermore, binding of Ras proteins to Raf kinase is not sufficient to activate Raf kinase: this process requires membrane or lipid components for reasons that we do not understand. A molecular description of these events may also lead to new therapeutic opportunities.
K-Ras 4B, the major isoform expressed in human cancers, has a function that is not shared by other members of the Ras family: it binds calmodulin. The consequences of this are currently being investigated, but this unique property might afford another way to attack this isoform selectively. Likewise, K-Ras 4B is localized in the plasma membrane through different mechanisms than other Ras proteins (a polybasic sequence at the C-terminus ensures plasma membrane localization, while other Ras proteins use palmitoylation for this purpose). A strategy that attempts to exploit this has been published recently Citation[7] and others are sure to follow.
4. Exploiting RNA interference
An alternative strategy for attacking K-Ras mutations directly involves use of RNA interference, coupled to specific delivery of siRNA to tumor cells. Successful application of this approach requires a safe and effective delivery system, with several key characteristics. The delivery system must be safe, obviously, and sufficiently stable to endure passage through the bloodstream. Its size and surface charge must be optimized to allow leakage from the vasculature system into the tumor bed. It needs to be internalized into tumor cells, through ligand-mediated endocytosis, for example. If particles accumulate in the endosomes, they need to escape into the cytoplasm to deliver their payload. The siRNA must be extremely potent, to allow maximum efficacy, to minimize off-target effects and to allow multiple siRNAs in the same payload. Targeted nanoparticles, such as those described by Davis and colleagues, appear to fulfill some of these criteria, resulting in cleavage of target mRNAs in humans for the first time Citation[8]. One remaining issue relating to K-Ras cancers is the choice of surface proteins to which nanoparticles attach and subsequently become internalized. Another is the precise payload. We have shown that systemic delivery of siRNA K-Ras to K-Ras tumor xenografts can slow down the growth of established tumors in mouse models, but addition of siRNA against PI3-kinase increases potency considerably, suggesting that the ideal therapeutic combination will consist of cocktail of siRNAs rather than siRNA against K-Ras alone. Similar results have been reported by Jacks, and coworkers Citation[9,10]. These studies represent early steps in a process that could eventually lead to a new platform for treating these and other cancers.
5. Harnessing the immune system
Perhaps the most exciting new approach to treating cancer involves harnessing the immune system to kill cancer cells, by relief of suppressive pathways that normally prevent successful attack. The first of these agents, anti-CTLA4 (cytotoxic T-lymphocyte-associated protein 4) and anti-PD1 (programmed cell death 1) have already been approved for treating metastatic or un-resectable melanoma, and clinical trials are underway in non-small and small cell lung cancer, among many other indications. Early results suggest this will be a successful approach in this disease Citation[11]. Furthermore, it seems likely that a better understanding of the mechanism involved in resistance and susceptibility to these agents will lead to significant improvements in response rates and safety. We can therefore expect, quite realistically, that these advances, in conjugation to the other approaches discussed above will soon lead to major advances in treating K-Ras lung cancers, as well as many other diseases that currently lack effective therapy.
6. Conclusion and future prospects
The urgency of the clinical need, the development of new technologies and insights and opportunities have converged to generate new interest in targeting Ras and Ras-driven cancers. This interest is apparent in academic labs, foundations, in the bio-pharmaceutical world and at the at the National Cancer Institute, from which the Ras National Program at Frederick National Laboratories for Cancer Research was launched. We hope to see these efforts translated into real clinical benefit over the next few years.
7. Expert opinion
Recent analysis from the The Cancer Genome Atlas consortium underscores the importance of the RTK-Ras pathway in lung adenocarcinoma: more than 70% of samples of cases can be accounted for by potential driver mutations in this pathway. The power of intervening in this pathway has been established by the success of small molecules that target mutant EGFR or ALK in these tumors. It is therefore reasonable to expect that compounds targeting K-Ras, the most frequent driver mutation in this disease, would have similar clinical impact. Targeting the MAPK or PI kinase pathways, both downstream of Ras, has been less successful so far, although many new combinations and approaches to attacking these pathways have yet to be tested. Targeting K-Ras directly therefore appears to be the most attractive small molecule approach to K-Ras driven cancers and recent breakthroughs in this area suggest this may indeed be possible, particularly for the most common form of oncogenic K-Ras in lung adenocarcinoma, the G12C K-Ras mutant.
While small molecule-based approaches now appear promising, they still face substantial technical challenges and, indeed, the approach of targeting K-Ras directly remains unproven. Other approaches are being developed that look equally promising. Of these, the possibility of harnessing the immune system is the most advanced and offers the greatest opportunity to impact this disease in the near future. This approach may be best suited to patients whose tumors have the highest mutational load and therefore the most neo-antigens for immune attack. How broadly this approach will be effective remains to be seen, but the prospect of long-term survival benefits is extremely exciting.
Another new technology that could impact this disease is based on the delivery of siRNA targeting K-Ras, or other targets, to these tumors. This technology offers the possibility of knocking down multiple targets simultaneously and has the power to be tailored to any combination of driver mutations that play causal roles in specific tumors. This technology also faces major technical challenges, but, as with small molecules and immune-based approaches, offers reasons to be optimistic for the future treatment of these diseases.
Declaration of interest
The author has no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Bibliography
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511(7511):543-50
- Arteaga CL, Engelman JA. ERBB receptors: from oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell 2014;25(3):282-303
- Stephen AG, Esposito D, Bagni RK, et al. Dragging ras back in the ring. Cancer Cell 2014;25(3):272-81
- Young A, McCormick F. Oncogenic and wild-type ras play divergent roles in the regulation of mitogen-activated protein kinase signaling. Cancer Discov 2013;3(1):112-23
- Ostrem JM, Peters U, Sos ML, et al. K-ras (G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 2014;503(7477):548-51
- Imielinski M, Berger AH, Hammerman PS, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 2014;150(6):1107-20
- Zimmerman G, Papke B, Ismail S, et al. Small molecule inhibition of the KRAS-PDEδ interaction impairs oncogenic KRAS signaling. Nature 2013;497(7451):638-42
- Davis ME, Zuckerman JE, Choi CH, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature 2010;454(7291):1067-70
- Yuan TL, Fellmann C, Lee CS, et al. Development of siRNA payloads to target KRAS-mutant cancer. Cancer Discovery 2014;4(10):1182-97
- Xue W, Dahlman JE, Tammela T, et al. Small RNA combination therapy for lung cancer. Proc Natl Acad Sci USA 2014;111(34):E3553-61
- Creelan BC. Update on immune checkpoint inhibitors in lung cancer. Cancer Contr 2014;21(1):80-9