Abstract
The increasing resistance of bacteria to conventional antimicrobial therapy within both the nosocomial and community environment has enforced the urgent requirement for the discovery of novel agents. This has stimulated increased research efforts within the field of lantibiotic discovery. Lantibiotics are ribosomally synthesised, post-translationally modified antimicrobial peptides that exhibit antimicrobial activity against a range of multi-drug-resistant (MDR) bacteria. The success of these agents as a novel treatment of MDR infections is exemplified by: the clinical development of MU1140 (mutacin 1140) and NAI-107 (microbisporicin), which are in late pre-clinical trials against gram-positive bacteria; NVB302 that has completed Phase I clinical trials for the treatment of Clostridium difficile infections and; duramycin that has completed Phase II clinical trials in the treatment of cystic fibrosis. Despite these potential successes, the traditional method of lantibiotic discovery involving the induction, production and identification is often an inefficient, time-consuming process creating a barrier to the efficient discovery of novel lantibiotics. The introduction of novel and innovative identification methods, including the application of probes and the ability to improve the stability and activity of agents via mutagenesis offer encouraging new areas to explore. The rapid expansion of available genome sequences of a wide variety of bacteria has revealed multiple interesting lantibiotic clusters that have the potential to be effective antimicrobials. However, due to the inefficient expression, screening and production methods currently employed, they are being assessed inefficiently and not rapidly enough to keep up with the ever-increasing demand for new agents.
1. Introduction
The emergence of new agents onto the market exhibiting novel mechanisms of action has been extremely limited over the past few decades, compounding the need to discover novel agents, particularly those with a novel mechanism of action as this is thought to decrease the potential of resistance development.
A report by the European centre for disease prevention states that of the 167 novel antimicrobial agents currently being assessed only 2 have a novel mechanism of action and only 2 new classes of agents have been marketed to treat multi-drug-resistant (MDR) gram-positive bacteria, the oxazolidinones and lipopeptides, since the 1970s.
This is alarming considering each year 25,000 patients die in the EU alone from MDR infections. The economic impact of these infections is also high with associated increased health-care costs and reduction in productivity costing the economy approximately EUR 1.5 billion per year Citation[1].
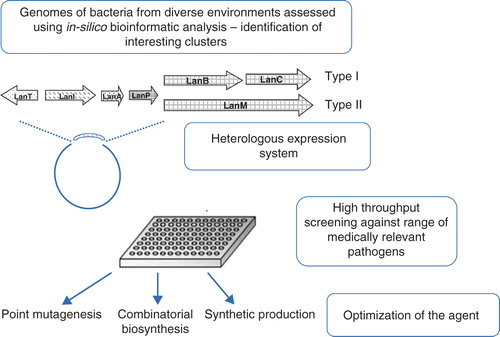
Lantibiotics are highly modified peptides containing unusual amino acids lanthionine and methyl-lanthionine that are divided into subclasses based on the type of structure they adopt and the biosynthetic enzymes involved in their production. The main classes include type I lantibiotics, which tend to be elongated, flexible structures that are modified by two enzymes encoding a dehydratase (LanB) and a cyclase (LanC). Type II lantibiotics form more globular, rigid structures and are modified by a single biosynthetic enzyme LanM performing both dehydratase and cyclase functions. A typical lantibiotic operon also contains genes for transport/processing (LanT), immunity (LanI), proteolytic processing (LanP) and structural gene (LanA) as shown in . Despite similarities between lantibiotic types, over 15 post-translational modifications have been described thus leading to a wide variety of structures Citation[2].
Figure 1. Schematic representation of a strategy that could be applied for a more streamlined protocol for the discovery of novel lantibiotics. The operon depicts the general organisation of genes involved in lantibiotic production.
The biosynthesis and lantibiotic ring structure were first described for lantibiotics epidermin and gallidermin Citation[3-5], leading to the term lantibiotics being adopted to describe these agents. One of the most attractive features of these agents is their multiple modes of action Citation[6,7], which often includes interaction with complex intermediates such as lipid II, a highly conserved molecule essential for cell wall synthesis, which is less prone to structural changes Citation[8,9]. Nisin, for example, targets the pyrophosphate in lipid II as opposed to glycopeptides that interact with the variable pentapeptide Citation[10]. The range of activity lantibiotics exhibit against MDR bacteria also makes them ideal candidates as novel agents against MDR infections Citation[11-14]. Although traditional methods have aided the isolation of some promising agents, one of the real failures regarding lantibiotic discovery is the current inability to fully investigate and exploit the numerous potentially interesting lantibiotic encoding clusters identified in a wide range of bacterial genomes.
2. Lantibiotics – a long road to discovery
Despite nisin being successfully used as a food additive to control food-borne pathogens for over the past 50 years Citation[15,16] and the recognition of lantibiotics potential against a range of pathogenic bacteria with the advancement of peptides MU1140 Citation[17,18], NAI-107 Citation[19], NVB302 Citation[20] and duramycin Citation[21] into pre-clinical and clinical trials, no lantibiotic is currently used within the clinical environment.
The discovery of lantibiotics has often involved a relatively long process relying on plate-based methods, including simultaneous and deferred antagonism and a range of heat treatment and enzyme characterisation studies Citation[22,23]. Their discovery is also hindered by the fact that the producing strains often produce the peptides in very low amounts, if at all. Production is often reliant upon finding the specific trigger for production, which can include testing a range of nutrient-rich and poor media at a variety pH ranges, including the addition or exclusion of specific elicitor agents Citation[24,25]. Quorum sensing systems have also been linked to lantibiotic production, for example the accessory gene regulator (agr) system in Staphylococci controls the extracellular processing of the leader peptide of epidermin Citation[26]. Lantibiotics nisin and mersacidin can also auto-induce their own production Citation[27,28]. To determine the specific factors that trigger lantibiotic production is therefore extremely time-consuming and does not guarantee success. When activity is observed further investigation is required for the exact identification of the agent responsible, which can potentially turn out to be a previously described agent. Various methods previously employed in lantibiotic discovery are highlighted in .
Table 1. Examples of the various methods utilised in the discovery of lantibiotics.
The development of techniques that could facilitate the rapid analysis and identification of novel lantibiotics is being developed; however, a cohesive combination of these methods is urgently required to make lantibiotic discovery a more efficient process.
When considering the detection of lantibiotics from bacterial cultures, the application of more rapid techniques including liquid chromatography coupled to ESI-mass spectrometry would be much more sensitive and productive when compared to traditional methods Citation[29,30]. This would be even more effective if an extensive database of mass spectrometry and NMR data existed for lantibiotics and would drastically reduce dereplication time.
Lantibiotic probes are an interesting and potentially useful method of lantibiotic discovery, these works by exploiting the presence of thioether rings that can be reversibly opened; however, they have only been applied to pure cultures of known lantibiotics and are yet to be fully investigated when considering their specificity and sensitivity in mixed microbial cultures. The effect the probes may have on bioactivity also requires investigation Citation[31,32].
3. Utilising genome data for the discovery of novel lantibiotics
One of the most promising areas of research regarding lantibiotic discovery is the application of knowledge gained through genome sequencing data. Specific web-based genome mining tools, including Bagel and antiSMASH, have been developed to aid the discovery of novel antimicrobial peptide gene clusters using bacteriocin and motif databases incorporating the analyses of surrounding genes. The biosynthetic enzymes involved in lantibiotic modification contain highly conserved domains, which can be utilised as driver sequences to screen genomic data for new lantibiotics. This method was applied in the discovery of novel lantibiotic linchenicidin, using genes encoding LanM proteins as a driver sequence to screen genomes of bacteria not previously recognised as lantibiotic producers Citation[33]. It is thought, however, that this reliance on homologous sequences will potentially only uncover those with similarity to known lantibiotics and greater interrogation of genomes is required. The in silico bioinformatic analysis of available genomes could also be applied to allow a more directed approach to the selection of specific environments to target for the discovery of novel lantibiotics Citation[34-36].
Despite advancements in genome sequencing and bioinformatics programs leading to the identification of unknown lantibiotic producing bacteria and interesting operons encoding lantibiotics, a robust heterologous expression system to enable their production has yet to be described. Traditional methods of lantibiotic production can result in manufacturing difficulties, including problems with up-scaling fermentations and batch-to-batch variations of agents, which can hinder the possibility of gaining FDA approval. The development of a robust, reproducible system for lantibiotic production could circumvent these issues and is essential for the discovery of novel lantibiotics as it allows the increased production of lantibiotics and also the efficient production of ‘silent’ or ‘sleeping’ lantibiotic clusters that may not be induced in the parent strain. The strain used for production also has to be carefully selected to ensure it is compatible with the expression of the various types of lantibiotic-producing operons.
Once an efficient heterologous system has been developed another major obstacle is the availability of a robust high-throughput screening system enabling interesting agents to be efficiently screened against a panel of medically relevant pathogens. Methods that could be applied include using a turbidometric assay Citation[37] or using a pH indicator to determine bacterial metabolism Citation[38]. High-throughput screening methods have proved effective in the discovery of lantibiotics NAI-107 Citation[39] and planosporicin Citation[40] and therefore warrant application in further studies.
In-vitro synthesis of lantibiotics is also an underexplored area of lantibiotic discovery, which includes semi-synthesis, using chemical modifications to increase diversity Citation[41,42].
Solid-phase peptide synthesis is one of the most promising techniques in the synthetic production of lantibiotics and has proved effective for the production of certain agents Citation[43,44]. However, for this method to be truly efficient the optimisation of reactions is required to allow increased yields and cost-effectiveness of this method. A synthetic analogue of the lantibiotic MU1140 (mutacin 1140), termed MU1140-S is currently in late pre-clinical trials. The company producing this agent, Oragenics, claim to have overcome the manufacturing difficulties associated with lantibiotics by application of its Differentially Protected Orthogonal Lanthionine Technology. It is believed cost-effective, large-scale and consistent production can be achieved using this method of synthesis, if so, this offers a real breakthrough in the ability to produce highly effective lantibiotics with consistent yields and purity.
4. The application of methods to improve known and novel lantibiotics
Various technologies are now available for the improvement of lantibiotics, including point mutations that have produced improved agents exhibiting increased stability, solubility and potency Citation[45,46]. This method has mainly been applied to reference agents and requires a robust high-throughput screening system to be in place to screen all the variants produced. Site-directed mutagenesis has also revealed the importance of specific amino acids and provided essential information to allow the rational design of lantibiotics with greater potency, stability and lower toxicity. The mode of action of lantibiotics often includes pore formation and specific interactions with cell wall precursors, including lipid II and lipid I Citation[47]. Increasing knowledge of specific structure–function relationships can highlight specific areas of interest. For example, site-directed mutagenesis of the hinge region of nisin Z which originally possessed activity exclusively against gram-positive bacteria resulted in gram-negative activity being developed Citation[45]. Extensive characterisation of lantibiotics is essential to facilitate their application within the medical environment.
Combinatorial biosynthesis is also a promising method for the rational design of novel lantibiotics Citation[48]; however, for this to be truly efficient a more in-depth knowledge of the actual impact the specific modifications can have on lantibiotics stability, solubility and activity is required. The combination of lantibiotics and known antimicrobial agents could also be investigated. This method has been successfully applied in the combination of nisin and vancomycin that when combined were active against vancomycin resistant enterococcus Citation[49] and therefore warrants further investigation.
Greater understanding is also required into the effect specific modifications have on the lantibiotics ability to interact with target pathogens and exert their antimicrobial effect. This could aide the development of tools to predict ‘in silico’ interactions of specific agents enabling only the most promising agents to be taken forward into the lab. highlights methods that could be applied for the streamlined discovery and engineering of novel lantibiotics.
In conclusion, the development of technologies that can be applied to the discovery of lantibiotics offers great potential in streamlining the process. However, these need to be refined before their benefit will be realised.
5. Expert opinion
Due to the constantly increasing rates of resistance developing against currently used antimicrobials, the search for novel agents with favourable activity against these resistant bacterial strains is essential.
The lantibiotics appear to be ideal candidates for the treatment of MDR infections owing to their activity against a range of MDR strains and multiple modes of action, which is thought to restrict the rate of resistance development towards these agents.
Despite the wealth of novel lantibiotic clusters in a wide range of bacterial strains that have been highlighted through genome sequencing, the actual discovery and identification of novel lantibiotics often relies on time-consuming traditional isolation and screening techniques. The success of this approach is determined by the ability to find the precise inducing conditions to stimulate lantibiotic production. Even when strains contain multiple lantibiotic clusters within their genome, there is no guarantee their production will be observed in the laboratory as they can often be present but are ‘silent’ gene clusters. Despite novel technologies that can be applied to the discovery of lantibiotics, they are not currently being applied at a rate that enables the efficient analysis of these agents and this must be addressed. There are two areas that novel technologies can be applied, one in the identification of agents from bacterial cultures and possibly the most promising area, from the analysis of bacterial genomes. Although technologies exist that can aide lantibiotic identification, there is currently no standard method of efficiently processing these samples and many of the technologies are in the early stage of development and require application to a greater number of samples and studies to identify and highlight there true potential and efficiency.
The ideal solution to this problem of processing data arising from genome analysis is for the development of a robust heterologous system that enables interesting gene clusters to be expressed under a strong constitutive promoter with the host strain optimised for production in specified culture conditions. This would eliminate the need to spend excessive time screening multiple different medium or elicitors to observe production and also make up-scaling production easier and increase batch-to-batch reproducibility. The success of this approach would be highly dependent upon the application of a high-throughput screening system that could be applied using panels of medically relevant pathogenic bacteria. Once a promising agent is identified site mutagenesis and combinatorial biosynthesis could then be applied to increase the stability, solubility and potency of the agent. However, for this approach to be efficient and create meaningful results further work is necessary to understand the impact of specific modifications on the agent and structure–activity relationships. Although the interesting methods of discovery and modifications including the use of probes, mutagenesis, combinatorial biosynthesis and semi and full synthesis, none of these methods have been extensively applied to a wide range of lantibiotics and are fully optimised for the high-throughput analysis of agents. Despite the clinical promise of lantibiotics and novel methods of discovery further work is required to streamline these methods enabling the accurate and efficient assessment of the vast array of lantibiotic clusters available.
Declaration of interest
The author has no relevant affiliation or financial involvement with any organisation or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Bibliography
- European centre for disease prevention and control/European medicine agency joint technical report. The bacterial challenge: Time to react. Press Release 2009. doi 10.2900/2518
- Oman TJ, van der Donk WA. Follow the leader: the use of leader peptides to guide natural product biosynthesis. Nat Chem Biol 2010;6(1):9-18
- Allgaier H, Jung G, Werner RG, et al. Epidermin: sequencing of a heterodet tetracyclic 21-peptide amide antibiotic. Eur J Biochem 1986;160:9-22
- Schnell N, Entian KD, Schneider U, et al. Prepeptide sequence of epidermin, a ribosomally synthesized antibiotic with four sulphide-rings. Nature 1988;333(6170):276-8
- Götz F, Perconti S, Popella P, et al. Epidermin and gallidermin: staphylococcal lantibiotics. Int J Med Microbiol 2014;304(1):63-71
- Payne DJ, Gwynn MN, Holmes DJ, et al. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Disc 2007;6(1):29-40
- Dischinger J, Chipalu SB, Bierbaum G. Lantibiotics: promising candidates for future applications in health care. Int J Med Microbiol 2014;304(1):51-62
- Pag U, Sahl HG. Multiple activities in lantibiotics - models for the design of novel antibiotics? Curr Pharm Des 2002;8(9):815-33
- Brötz H, Josten M, Wiedemann I, et al. Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol Microbiol 1998;30(2):317-27
- Hasper HE, Kramer NE, Smith JL, et al. An alternative bactericidal mechanism of action for lantibiotic peptides that target lipid ii. Science 2006;313(5793):1636-7
- Boakes S, Wadman S. The therapeutic potential of lantibiotics. Drug Dis Devel 2008;27:22-5
- Nascimento JS, Ceotto H, Nascimento SB, et al. Bacteriocins as alternative agents for control of multiresistant staphylococcal strains. Lett Appl Microbiol 2006;42:215-21
- Fontana MB, Bastos MC, Brandelli A. Bacteriocins pep5 and epidermin inhibit staphylococcus epidermidis adhesion to catheters. Curr Microbiol 2006;52:350-3
- Kruszewska D, Sahl HG, Bierbaum G, et al. Mersacidin eradicates methicillin-resistant staphylococcus aureus (mrsa) in a mouse rhinitis model. J Antimicrob Chemother 2004;54(3):648-53
- Lubelski J, Rink R, Khusainov R, et al. Biosynthesis, immunity, regulation, mode of action and engineering of the model lantibiotic nisin. Cell Mol Life Sci 2008;65(3):455-76
- Mattick AT, Hirsch A, Berridge NJ. Further observations on an inhibitory substance (nisin) from lactic streptococci. Lancet 1947;250(6462):5-8
- Ghobrial O, Derendorf H, Hillman JD. Pharmacokinetic and pharmacodynamic evaluation of the lantibiotic mu1140. J Pharm Sci 2010;99(5):2521-8
- Ghobrial OG, Derendorf H, Hillman JD. Pharmacodynamic activity of the lantibiotic mu1140. Int J Antimicrob Agents 2009;33(1):70-4
- Jabes D, Brunati C, Candiani G, et al. Efficacy of the new lantibiotic nai-107 in experimental infections induced by multidrug-resistant gram-positive pathogens. Antimicrob Agents Chemother 2011;55(4):1671-6
- Crowther GS, Baines SD, Todhunter SL, et al. Evaluation of nvb302 versus vancomycin activity in an in vitro human gut model of clostridium difficile infection. J Antimicrob Chemother 2013;68(1):168-76
- Jones A, Helm J. Emerging treatments in cystic fibrosis. Drugs 2009;69(14):1903-10
- Tagg J, Dajani A, Wannamaker L. Bacteriocins of gram-positive bacteria. Bacteriol Rev 1976;40(3):722-56
- Tagg JR, Bannister LV. “Fingerprinting” beta-haemolytic streptococci by their production of and sensitivity to bacteriocine-like inhibitors. J Med Microbiol 1979;12(4):397-411
- Jack RW, Tagg JR, Ray B. Bacteriocins of gram-positive bacteria. Microbiol Rev 1995;59:171-200
- Zhu H, Sandiford S, Wezel G. Triggers and cues that activate antibiotic production by actinomycetes. J Ind Microbiol Biotechnol 2014;41(2):371-86
- Kies S, Vuong C, Hille M, et al. Control of antimicrobial peptide synthesis by the agr quorum sensing system in staphylococcus epidermidis: activity of the lantibiotic epidermin is regulated at the level of precursor peptide processing. Peptides 2003;24:329-38
- Kuipers OP, Beerthuyzen MM, de Ruyter PG, et al. Autoregulation of nisin biosynthesis in lactococcus lactis by signal transduction. J Biol Chem 1995;270(45):27299-304
- Schmitz S, Hoffmann A, Szekat C, et al. The lantibiotic mersacidin is an autoinducing peptide. Appl Environ Microbiol 2006;72(11):7270-7
- Perez R, Zendo T, Sonomoto K. Novel bacteriocins from lactic acid bacteria (lab): various structures and applications. Microb Cell Fact 2014;13(Suppl 1):S3
- Zendo T, Nakayama J, Fujita K, et al. Bacteriocin detection by liquid chromatography/mass spectrometry for rapid identification. J Appl Microbiol 2008;104(2):499-507
- Li J, Girard G, Florea BI, et al. Identification and isolation of lantibiotics from culture: a bioorthogonal chemistry approach. Org Biomol Chem 2012;10(43):8677-83
- Smith L, Novák J, Rocca J, et al. Covalent structure of mutacin 1140 and a novel method for the rapid identification of lantibiotics. Eur J Biochem 2000;267(23):6810-16
- Begley M, Cotter PD, Hill C, et al. Identification of a novel two-peptide lantibiotic, lichenicidin, following rational genome mining for lanM proteins. Appl Environ Microbiol 2009;75(17):5451-60
- van Heel AJ, de Jong A, Montalbán-López M, et al. Bagel3: Automated identification of genes encoding bacteriocins and (non-)bactericidal posttranslationally modified peptides. Nucleic Acids Res 2013;41(W1):W448-53
- Blin K, Medema MH, Kazempour D, et al. Antismash 2.0–a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res 2013;41(W1):W204-12
- Hammami R, Zouhir A, Le Lay C, et al. Bactibase second release: a database and tool platform for bacteriocin characterization. BMC Microbiol 2010;10(1):22
- De La Fuente R, Sonawane ND, Arumainayagam D, et al. Small molecules with antimicrobial activity against e. Coli and p. Aeruginosa identified by high-throughput screening. Br J Pharmacol 2006;149(5):551-9
- Ymele-Leki P, Cao S, Sharp J, et al. A high-throughput screen identifies a new natural product with broad-spectrum antibacterial activity. PLoS One 2012;7(2):e31307
- Castiglione F, Lazzarini A, Carrano L, et al. Determining the structure and mode of action of microbisporicin, a potent lantibiotic active against multiresistant pathogens. Chem Biol 2008;15(1):22-31
- Castiglione F, Cavaletti L, Losi D, et al. A novel lantibiotic acting on bacterial cell wall synthesis produced by the uncommon actinomycete planomonospora sp. Biochemistry 2007;46(20):5884-95
- Rollema HS, Kuipers OP, Both P, et al. Improvement of solubility and stability of the antimicrobial peptide nisin by protein engineering. Appl Environ Microbiol 1995;61(8):2873-8
- Field D, Begley M, O’Connor PM, et al. Bioengineered nisin a derivatives with enhanced activity against both gram positive and gram negative pathogens. PLoS One 2012;7(10):e46884
- Mothia B, Appleyard AN, Wadman S, et al. Synthesis of peptides containing overlapping lanthionine bridges on the solid phase: an analogue of rings d and e of the lantibiotic nisin. Org Lett 2011;13(16):4216-19
- Liu W, Chan AS, Liu H, et al. Solid supported chemical syntheses of both components of the lantibiotic lacticin 3147. J Am Chem Soc 2011;133(36):14216-19
- Yuan J, Zhang ZZ, Chen XZ, et al. Site directed mutagenesis of the hinge region of nisin z and properties of nisin z mutants. Appl Microbiol Biotechnol 2004;64:806-15
- Appleyard AN, Choi S, Read DM, et al. Dissecting structural and functional diversity of the lantibiotic mersacidin. Chem Biol 2009;16(5):490-8
- Bonelli RR, Schneider T, Sahl HG, et al. Insights into in vivo activities of lantibiotics from gallidermin and epidermin mode of action studies. Antimicrob Agents Chemother 2006;50:1449-57
- van Heel AJ, Mu D, Montalbán-López M, et al. Designing and producing modified, new-to-nature peptides with antimicrobial activity by use of a combination of various lantibiotic modification enzymes. ACS Synthetic Biology 2013;2(7):397-404
- Arnusch CJ, Bonvin AM, Verel AM, et al. The vancomycin-nisin(1-12) hybrid restores activity against vancomycin resistant enterococci. Biochemistry 2008;47(48):12661-3
- Whitehead HR. A substance inhibiting bacterial growth, produced by certain strains of lactic streptococci. Biochem J 1933;27(6):1793-800
- Jansen EF, Hirschmann DJ. Subtilin. An antibacterial product of bacillus subtilis. Culturing conditions and properties. Arch Biochem Biophys 1944;4:297-309
- Fuchs SW, Jaskolla T, Bochmann S, et al. Entianin, a novel subtilin-like lantibiotic from bacillus subtilis subsp. Spizizenii dsm 15029t with high antimicrobial activity. Appl Environ Microbiol 2011;77(5):1698-707
- Hillman JD, Novák J, Sagura E, et al. Genetic and biochemical analysis of mutacin 1140, a lantibiotic from streptococcus mutans. Infect Immun 1998;66(6):2743-9
- Georgalaki MD, Van den Berghe E, Kritikos D, et al. Macedocin, a food-grade lantibiotic produced by streptococcus macedonicus aca-dc 198. Appl Environ Microbiol 2002;68(12):5891-903
- Ross KF, Ronson CW, Tagg JR. Isolation and characterization of the lantibiotic salivaricin a and its structural gene sala from streptococcus salivarius 20p3. Appl Environ Microbiol 1993;59(7):2014-21
- Lawton EM, Cotter PD, Hill C, et al. Identification of a novel two-peptide lantibiotic, haloduracin, produced by the alkaliphile bacillus halodurans c-125. FEMS Microbiol Lett 2007;267(1):64-71

