Abstract
Background
Oncogenic viruses, their possible association with breast cancer (BC) and effect on its clinical course are interesting issue. The present study evaluates the presence of human papillomavirus (HPV), EpsteinBarr virus (EBV), and human mammary tumor virus (HMTV) in BC and their relation with clinico-pathological characteristics.
Patients and Methods
This study was conducted on 80 Egyptian women with BC and 30 control women without known oncological disease. Forty formalin-fixed paraffin-embedded (FFPE) tissues, forty fresh tissue samples, and white blood cells (WBCs) of BC patients and WBCs of controls were subjected to a qualitative polymerase chain reaction (PCR). Quantitative real-time PCR was used to measure viral loads in fresh tissues of BC. The result was correlated with clinico-pathological characteristics of BC.
Results
HPV was detected in 33 (41.25%), EBV in 30 (37.5%) and HMTV in 33 (41.25%) BC patients. None of the control women was positive for HPV or EBV while HMTV was detected in 7 (23.3%). Among 40 BC WBCs specimens, HPV/HMTV were found together in 25%, followed by EBV/HMTV in 2.5% and EBV/HPV in 2.5%. However, the three viruses (HPV/EBV/HMTV) were found together in only 5%. In the 40 fresh BC tissues, the three viruses were found together in 12 (30%), EBV/HMTV in 7 (17.5%), HPV/HMTV in 4 (10%), and HPV/EBV in 4 (10%). EBV, HMTV, or multiple viral infections were associated with younger age of BC women. HPV, EBV, and HMTV median loads in fresh tissues were 4.8×103 copies/μL, 6.3×103 copies/μL, and 97 copies/μL, respectively.
Conclusion
WBCs could be a more suitable specimen instead of fresh tissue for HMTV detection in BC patients to avoid invasive procedures. The presence of HPV, EBV, and HMTV together in Egyptian women with BC was significantly associated with younger age.
Introduction
New breast cancer (BC) cases among Egyptian females in 2020 were 22,038, representing 32.4% among other cancer types with 10.3% deaths.Citation1 Human Papillomavirus (HPV), EpsteinBarr virus (EBV), and Human Mammary Tumor virus (HMTV) were suggested to have a possible relationship to BC.Citation2,Citation3 Simões and co-workersCitation4 summarized 29 studies including 2211 breast tissue samples from different geographical regions. They found that 23% of BC patients had HPV DNA. Besides, HMTV was reported in a wide range (0–78%) of numerous populations with BCs.Citation5 Prevalence range of EBV infection among BC cases of 26 countries was 0–78.12%.Citation6
Individuals with BC may become immune-compromised due to BC disease itself or chemotherapy and radiotherapy. Hence, they might be more vulnerable to viral infections that can aggravate the disease.Citation7 For example, HPV Early 6/Early 7 (E6/E7) oncoproteins can convert non-invasive and non-metastatic BC cells into invasive and metastatic forms.Citation8 Also, the presence of HPV in BC patients was associated with increased inflammatory cytokines, tumor progression,Citation9 and younger age.Citation10 HMTV showed higher positivity in patients with advanced (grade 3) BC diseaseCitation11 and was correlated with estrogen and human epidermal growth factor receptor 2 (her2/neu) expression and metastasis in these patients.Citation12 Many studies have reported an association of EBV with lymph node metastasis and more aggressive behavior of the tumor.Citation13–Citation15 Moreover, HPV, EBV, and MMTV presence was previously reported to be associated with increased-grade in human BCs.Citation16
It was previously reported that HMTV could be detected together with HPV and EBV in the same BC cells.Citation16 Therefore, HPV, EBV, and HMTV alone or together in the same BC patient were highly suggested to cause BC disease aggressiveness. Limited reports from the Arab world on the co-prevalence of HPV, EBV, and HMTV and their clinical relevance in BC patients were found. The different reviewed literature included HPV and EBV together. Still, they did not investigate their co-prevalence with HMTV or their association with clinico-pathological characteristics of BC disease, such as studies conducted on Syrian BC womenCitation17 and Egyptian BC women.Citation18,Citation19 In addition, another study detected HPV, EBV, and HMTV in Egyptian BC women, but it did not investigate their association with clinico-pathological characteristics of BC disease.Citation20
BC disease management and understanding its behavior requires extensive study of the association between environmental factors that may affect the pathogenesis of BC disease (especially viral infection) and their association with patient’s clinico-pathological parameters. The current study aimed to evaluate the frequency of HPV, EBV, and HMTV alone or together in different samples of BC disease and their association with some clinical and pathological characteristics as a possible indicator of BC patients at a higher risk to develop severe disease.
Patients and Methods
Patients
This study was conducted on 80 BC patients diagnosed and treated at the medical oncology department, National Cancer Institute (NCI), Cairo University, between July 2018 and June 2020. Thirty normal control women without known oncological disease and with comparable age were included as a control group. The Institutional Review Board (IRB) of the NCI approved the protocol IRP NO. (201819039.3). Informed consent was taken from all participants who joined the study. Women suffering from BC and who had not received any chemotherapy treatment before surgery were the inclusion criteria.
Collection of Patients’ Data
The patients’ data were collected from their medical files kept in the biostatistics and epidemiology department, NCI, Cairo University. Data extracted were age at diagnosis, family history of BC, diagnosis, lymph node metastasis, tumor size, histologic grade, laterality, hormonal receptor status including estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2). Histologic type and histological grade were evaluated consistently with the previous protocols.Citation21,Citation22
Specimen Collection
Five mL of whole blood and 250 mg of fresh tissues were collected from 40 BC patients. In addition, 250 mg of formalin-fixed and paraffin-embedded (FFPE) tissue were collected from another 40 BC women. Five mL of whole blood were obtained from 30 controls. Tissue analysis was limited to core biopsies only. White blood cells (WBCs) were separated from blood samples according to the previously published protocolsCitation23,Citation24 while processing, tissues, and WBCs were stored at −80°C.
Nucleic Acid Extraction
Viral DNAs were extracted from WBCs using QIAamp viral RNA extraction kit (Qiagen, Valencia, USA), which can extract both viral DNAs and RNAs. Genomic DNAs were extracted from fresh and FFPE tissues using Gene JET Genomic DNA Purification Kit (Thermo Scientific, Inc, USA) and QIAamp DNA FFPE Tissue Kit (Qiagen, Valencia, USA), respectively, according to the manufacturer’s instructions. A Nano-Drop 2000 spectrophotometer (Thermo Scientific/US, Canada) was used to assess the quality and quantity of isolated viral and total genomic DNAs. 100 ng of DNA templates were used in each Polymerase chain reaction (PCR) assay. DNA extracts were stored at −80 °C till further analysis.
Molecular Detection of Viral DNAs
Every feasible precaution was taken to avoid contamination of the analyzed samples whether in qualitative or quantitative PCR assays. These precautions included work in separate areas with a one-way direction using appropriate personal protective equipment (PPE) for; sample processing, pre-amplification, and post-amplification. Besides, all the used equipment and surfaces were continuously de-contaminated, all samples were stored separately from kits and reagents. Also, precautions like dispensing reagents into aliquots, using pipettes with barrier tips, and changing gloves before dealing with any of the samples, were implemented. Finally, the result was validated and reported after validating positive and negative controls.
Qualitative PCR
All extracted DNAs from patients (FFPE tissues, fresh tissues and WBCs) and controls (WBCs) were subjected to qualitative conventional PCR assay targeting HPV Late 1 (L1) region detection, EBV Bam H1W region detection, and HMTV envelope (env) gene detection. The amplification was conducted in a final volume of 25 μL of 1X AmpliTaq PCR master mix (Biosystems, Barcelona, Spain) and 0.2 μM of both forward and reverse primers for each virus, using thermal cycling block (Applied Biosystems™ ProFlex™ PCR System, USA). Nuclease-free water was used to make up the volume. Primers used in the molecular assays are presented in . The PCR reaction amplification conditions for HPV, EBV, and HMTV were carried out according to previously published protocols.Citation25–Citation28
Table 1 Sequences of Primers Used for Human Papillomavirus (HPV), EpsteinBarr Virus (EBV) and Human Mammary Tumor Virus (HMTV) by Both Qualitative and Quantitative Polymerase Chain Reaction (PCR)
PCR reaction mixture without template DNA was used as a negative control sample in each PCR run. EBV (American Type Culture Collection (ATCC®) VR-1492™) was used as a positive control for EBV. HPV & HMTV DNAs were extracted in NCI virology laboratory from Michigan Cancer Foundation-7 (MCF7) cellsCitation28–Citation30 (ATCC® HTB-22™) obtained from Egyptian Holding Company for Biological Products & Vaccines (VACSERA) using the same extraction procedure for samples. HPV positive control was confirmed as HPV type 18 by PCR using a linear array HPV genotyping kit (Roche Diagnostics, Indianapolis, IN, USA).
Seventeen μL of each PCR product were electrophoresed on a 2% agarose gel (Sigma) in Tris-acetate buffer (TAE 1X) and then stained with 0.5 μg/mL ethidium bromide, examined under Ultra Violet (UV) transillumination and photographed. The product sizes were compared against 100 bp DNA ladder (Genedirex, Taiwan).
Specificity of qualitative PCR for detection of HPV, EBV, and HMTV DNAs was performed by testing samples positive for the following viruses; Herpes Simplex Virus 1 (HSV1), Herpes Simplex Virus 2 (HSV2), Adenovirus (Adv.), HMTV, HPV, Cytomegalovirus (CMV) and EBV, according to the tested virus. Then, qualitative PCR assay was performed according to the previously published protocols.Citation25–Citation28
Quantitative Real-Time PCR
HPV, EBV, and HMTV viral loads were measured in fresh tissues by a quantitative real-time detection system (Applied Biosystems 7500 Fast Real-Time PCR system Thermal Cycling Block, USA). The amplification was conducted in a final volume of 25 μL containing 1X Qiagen Sybrgreen® PCR Master Mix kit (Qiagen, Valencia, USA), 0.2 μM of each forward and reverse primer for each virus, and 100ng of extracted DNA. Nuclease-free water was used to make up the volume. Thermal cycling was performed as mentioned before in qualitative PCR assay.
Threshold cycle (Ct) value was detected for each sample, and viral load was calculated. Post amplification melting temperature (Tm) analysis, together with gel electrophoresis, clearly differentiated nonspecific PCR products from the HPV, EBV, or HMTV specific product sequences. Standard curves were constructed using 10-fold serial dilutions (10 to 106 copies/μL) of HPV, EBV, or HMTV positive controls with known concentrations of 300 FG of extracted DNA equivalent to one copy of viral DNA.
Statistical Methods
Sample size calculation: the total number of cases that attended NCI with BC was estimated to be 2500 per year. A previous study conducted in Egypt by El-Sheshtawy et al (2017)Citation31 indicated that the overall prevalence of HPV in malignant breast tissue was 16.7%. For a two-sided 95% confidence interval for a single proportion based on these criteria, the minimum required sample size to evaluate the practice was 54 cases with a 10% margin of error. To compensate for possible loss, the sample increased by 10%. Therefore, a sample of 60 cases was estimated. The sample size was calculated using the n-Query statistical package.Citation32
Statistical analysis was done using IBM SPSS® Statistics version 22 (IBM® Corp., Armonk, NY, USA). Qualitative data were expressed as frequency and percentage. Pearson’s Chi-square test or Fisher’s exact test was used for examining the relationship between qualitative variables. Kappa test was used to evaluate the agreement of detection of viruses in fresh tissues and WBCs. Numerical data were expressed as mean and standard deviation or median and range. Comparison between two groups for numerical variables was made using either the Student-t-test or Mann–Whitney test according to distribution. A p-value< 0.05 was considered significant.
Result
Clinico-Pathological Findings
All studied BC patients (n=80) had a unilateral BC. Out of eighty BC patients, 34 (42.5%) were familial BC, including 24 BC patients with blood samples. Regarding BC disease characteristics, all BCs were diagnosed as invasive carcinoma (lobular or ductal), and 75/80 (93.8%) were classified as grade II tumors. The median tumor size was 2.5 cm, and the most observed was positivity for ER and PR hormonal receptors expression as observed in 70/80 (87.5%) and 72/80 (90%) of BC patients, respectively. Lymph node metastasis was found in 47/80 (58.8%) of patients. All pathological findings of BC patients are summarized in .
Table 2 Pathological Findings of Breast Cancer (BC) Patients
The mean age of the 80 BC patients was 41.3±9.2 years and 39.9±11.2 for the healthy controls, P=0.207. The forty BC women (with blood samples) and 30 control women were age-matched with no statistical difference (P=0.591). Besides, the two subgroups (FFPE and fresh tissue samples) of BC patients were comparable in age with a mean (38.6±6.7 years vs 41.3±9.2, respectively, P=0.140).
Presence of HPV, EBV and HMTV DNAs in Studied Groups as Detected by Qualitative PCR Assay
Taking all 80 BC women, HPV and HMTV were the most frequent viruses detected in BC specimens (33/80, 41.25%), followed by EBV (30/80, 37.5%) (Table S1). The detection of HPV, EBV, and HMTV DNAs, was significantly lower in FFPE tissues than in fresh tissue samples of BC patients. Therefore, data of FFPE tissues were not included in further analysis. Data are demonstrated in .
Table 3 HPV, EBV, and HMTV Frequency in FFPE Tissue and Fresh Tissue Samples of 80 BC Patients
Regarding WBCs of BC patients and controls, HPV, EBV, and HMTV DNAs were detected in 40%, 10%, and 57.5% of BC patients, respectively. Only HMTV was detected in 23.3% of healthy controls. Comparison of presence of viruses between BC and healthy control women showed P values of <0.001 and 0.004 for HPV and HMTV, respectively (Table S2).
Agreement of Viral Detection Between Fresh Tissues and Corresponding WBCs of 40 BC Women
The presence of HPV, EBV, and HMTV DNAs in fresh tissue alone, WBCs alone, or in both fresh tissues and WBCs, were analyzed to investigate the agreement between these two sample types (). HMTV was present in both fresh tissues and their corresponding WBCs of 21 (52.5%) BC patients with a moderate statistical agreement between fresh tissue and WBCs (Kappa=0.521, P= 0.001). However, there was no statistical agreement between fresh tissue and corresponding WBCs for the presence of HPV or EBV.
Table 4 Agreement of Viral Detection Between Fresh Tissues and Corresponding WBCs of 40 BC Women
Prevalence of Single and Multiple Infections with HPV, EBV, and HMTV in Fresh Tissue and WBCs Samples of 40 BC Women
According to qualitative PCR assay, patients were divided into two groups; patients positive for single viral DNA (HPV, EBV, or HMTV) and patients positive for 2 or 3 viral DNAs.
Regarding fresh tissues, single viral DNA was detected in 5 (12.5%) BC patients, while multiple infections were detected in 27 (67.5%) of the same group of patients. Regarding WBCs, single viral DNA was detected in 13 (32.5%) BC patients, while multiple infections were detected in 14 (35%) of the same group of patients.
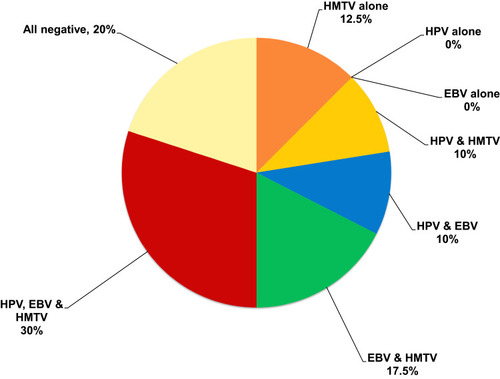
The most frequent multiple infections present in the fresh tissues of BC patients were HPV/EBV/HMTV together found in 12 (30%) out of 40 patients, followed by EBV/HMTV together in 7 (17.5%) patients. Moreover, HPV/HMTV DNAs together were present in 4 (10%) patients and HPV/EBV in 4 (10%) patients. HMTV DNA was the only virus present alone in 5 (12.5%) BC fresh tissues, and no HPV or EBV existed alone in any of the fresh tissue samples, as shown in .
Figure 1 Detection of single and multiple HPV, EBV and HMTV infection among 40 BC fresh tissue samples. HPV/EBV/HMTV together found in 30%, followed by EBV/HMTV in 17.5%, HPV/HMTV in 10%, and HPV/EBV in 10%. HMTV DNA was the only virus present alone in 12.5% of BC fresh tissues.
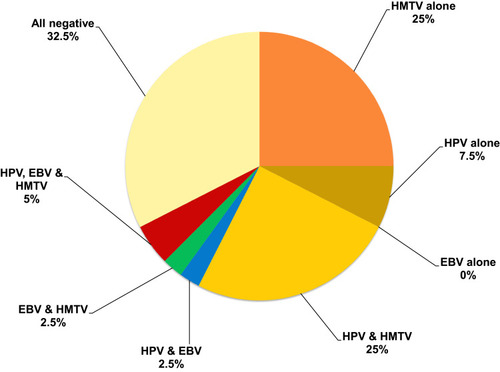
Regarding the presence of multiple viruses in WBCs of BC patients, HPV/EBV/HMTV DNAs together were found in only 2/40 (5%). The most frequent viruses that were present in co-infection, were HPV/HMTV (n=10, 25%) followed by EBV/HMTV (n=1, 2.5%) and EBV/HPV (n=1, 2.5%). HMTV was the predominating single virus (n=10, 25%) and then HPV (n= 3, 7.5%). No EBV existed alone in any of the WBCs of studied BC patients, as shown in .
Figure 2 Detection of single and multiple HPV, EBV and HMTV infection among 40 BC WBCs samples. HPV/EBV/HMTV DNAs together were found in only 5%. The most frequent viruses present in co-infection were HPV/HMTV in 25% followed by EBV/HMTV in 2.5% and EBV/HPV in 2.5%. HMTV was the predominating single virus present in 25% followed by HPV in 7.5%.
Specificity of HPV, EBV, and HMTV Primer Sequences Used for Qualitative PCR Assay
Such assays were found to be highly specific for the HPV L1 region, EBV Bam H1 W region, and HMTV env sequence as none of the other tested viruses was amplified.
Relations Between Presence of Viral DNAs and Some Clinico-Pathological Parameters of 40 BC Patients with Blood Samples
The statistical analysis showed a significant association between the high prevalence of EBV and HMTV DNAs among BC patients aged (≤45 years), P=0.015 and 0.001, respectively. Moreover, there was a significant association between the absence of Her2-neu receptor expression and the presence of HMTV DNA (P=0.018), as presented in . Also, the presence of multiple infections was statistically associated with younger age (≤45 years) of BC patients (P=0.014), as presented in .
Table 5 Relation Between Presence of HPV, EBV or HMTV Qualitative PCR and Clinico-Pathological Findings of 40 BC Patients
Table 6 Relation Between Co-Existence of Viruses and Clinico-Pathological Findings of 40 BC Patients
HPV, EBV, and HMTV DNAs Viral Loads
HPV, EBV, and HMTV DNA viral loads were detected by quantitative real-time PCR in fresh tissue samples obtained from BC patients that were positive by qualitative PCR assay. HPV median load was 4800 copies/μL ranged from 3.3×102 to 7×104 copies/μL. EBV median load was 6.3×103 copies/μL ranged from 49.8 to 120×104 copies/μL. In addition, HMTV median load was 97 copies/μL ranged from 1.2×101 to 1.3×103 copies/μL.
There was no significant association between EBV or HMTV viral load and any of clinico-pathological findings. However, HPV DNA viral load was significantly higher in patients ≤45 years (median: 1.2×104 copies/μL, range: 1.7×103 to 7×104 copies/µL) than older patients (median: 4.8 x102, range: 3.3×102 to 3.3×103 copies/μL, p <0.001).
Discussion
It was reported that some oncogenic viruses play a major role in the development of BC, and also their presence may aggravate disease outcomes.Citation7 HPV, EBV, and HMTV were the most investigated in many countries.Citation9,Citation12,Citation16,Citation19,Citation33 However, very limited studies investigated the presence of EBV with HPV and HMTV as single or multiple viruses and their association with clinico-pathological characteristics of BC disease.
The current study showed that HPV DNA was detected in 17.5% of FFPE tissue specimens, 50% of fresh BC tissues, and 40% of WBCs. This was in agreement with a previous report on Iranian BC women showing HPV DNA in 25.9% FFPE tissue specimen using PCR targeting the HPV L1 region.Citation35 Another study reported the presence of HPV DNA in fresh tissue of 48.6% of Iranian BC patients.Citation9 However, a report from Egypt demonstrated a higher percentage of HPV-16 in fresh tissues of 75.8% of non-inflammatory BC patients and 65.9% of inflammatory BC cases using PCR targeting HPV L1 region.Citation19 This difference between reports may be attributed to the difference in the pathology of the disease and the presence of a specific type of HPV.Citation36
In contrast, other reports showed a negative result for the presence of HPV DNA in FFPE BC tissues of 300 Iranian BC women,Citation37 81 Swiss BC females,Citation38 and 50 French BC females.Citation39 Another report from Egypt demonstrated the presence of HPV DNA in 16.7% of fresh BC tissues.Citation31 Other studies demonstrated the absence of HPV DNA in fresh tissues.Citation40 The difference in result may be due to low viral load,Citation41,Citation42 possible inter-laboratory inconsistency in collecting and handling samplesCitation38 and storage of specimens.Citation43
EBV DNA was detected in 17.5% of FFPE tissues, 57.5% of fresh tissues, and 10% of WBCs. A study from Egypt reported the detection of EBV DNA using PCR targeting EBV nuclear antigen-1 (EBNA-1) gene in 20% of FFPE BC tissues.Citation14 Moreover, it was detected in fresh samples of 53.3% of Sudanese BC patients using methylation-based PCR targeting Latent membrane protein-1 (LMP-1).Citation44 In agreement, a study performed by Marro et al (2014)Citation45 observed the presence of EBV DNA in 25.8% of FFPE BC tissues, but they reported higher result in peripheral blood mononuclear cell (PBMC) samples of 47% BC patients using quantitative real-time PCR of BXLF1 gene. In addition, a study from China reported EBV DNA in 24.4% of PBMCs of BC patients using Real-time PCR targeting BamH1-K.Citation46
Higher result of EBV DNA detection was also reported in 45% of Egyptian and 51.85% of Syrian FFPE BC tissues using PCR targeting EBNA-1 and LMP-1.Citation47,Citation48 Such discrepancy may be due to population characteristics differences,Citation49 decreased socioeconomic status of the infected persons,Citation50 difference in EBV derived nucleic acid target genes investigatedCitation51 and PCR false-positive result caused by amplicon contaminationCitation52 or presence of EBV in lymphocytes.Citation53
In contrast, a low percentage of EBV DNA was also found in 5.12% of FFPE tissues of Iranian women.Citation54 However, EBV DNA was not detected in FFPE tissues of Iranian women using PCR targeting the Bam HI-H rightward open reading frame 1 (BHRF1) region.Citation55 Moreover, EBV DNA was detected using PCR in 20% of fresh tissues of Egyptian BC patients,Citation56 4.7% of Mexican BC patients,Citation57 and 31% of Argentinian BC patients.Citation58 These lower detection rates could be due to differences in storage methods of samples.Citation59 At the same time, the false-negative result may be caused by 1) the presence of viral targets at levels that extend the sensitivity threshold of the assay,Citation13,Citation60 2) removal of Epstein–Barr virus-encoded small RNAs (EBER) coding sequences from the EBV genome,Citation50 3) alterations related to the integration of virus DNA into the host chromosomal DNA,Citation61 4) hit-and-run mechanism,Citation62 5) heterogeneous distribution of EBV genomes in morphologically matched tumor cells.Citation59
HMTV DNA was detected in 7.5% of FFPE tissues, 70% of fresh tissues, and 57.5% of WBCs of BC patients. In Saudi Arabia, Dossary and Associates (2018)Citation11 found viral DNA in 7.7% of FFPE BC tissue samples. Glenn et al (2012)Citation16 reported its presence in 78% of fresh DNA extracts from Australian BC women. This was in contrast with previous reports where HMTV DNA was present in 36% of FFPE tissues from BC Egyptians,Citation63 57.89% from BC Iraqis,Citation34 %32.2 from BC Iranians,Citation64 and 37.3% from BC Australians.Citation65 Even the picture was different when fresh tissues were used, where the lower result was also reported in many countries, including 13.9% in Tunisia,Citation66 37% in New York,Citation67 and 0% in Mexico.Citation57 In addition, MMTV-Long terminal repeat (LTR) gene sequence was detected in peripheral blood samples of 38% of BC patients from Pakistan.Citation12 Moreover, 300 WBCs samples of Iranian BC women were negative for MMTV.Citation68 These lower rates could be due to the probable low viral load in BCs, the extraction methods, and the sensitivity of PCR.Citation64,Citation65,Citation69,Citation70 Also, it was reported that mutations or rearrangements of viral sequences make their detection difficult.Citation66 Moreover, the high frequency of HMTV DNA in fresh tissue obtained in this study compared to studies from different countries may be due to a high level of contact with domestic animals.Citation71
Our result showed that the presence of viral DNAs in FFPE tissues was significantly lower than that in fresh tissue samples, possibly due to DNA degradation during paraffin fixation.Citation72 Therefore, we focused on the result obtained from fresh tissues for virological and clinical analysis. Interestingly, our result demonstrated a statistical agreement between fresh tissues and WBCs for the detection of HMTV in BC samples. This indicates a clinical relevance of WBCs as a suitable specimen instead of fresh tissue for HMTV detection in BC patients hence avoiding invasive procedures.
HPV and EBV DNAs were not detected in WBCs of healthy controls. This was in contrast to other studies which reported various percentages based on genetic, environmental, and geographical variations.Citation36,Citation46,Citation73–Citation75 However, HMTV DNA was detected in 23% of WBCs of our controls, possibly due to contact with their relative BC patients or exposure to the virus through vectors and environmental conditions.
The result showed that the presence of multiple viral infections was more frequent (67.5%) than the presence of a single infection (12.5%). Multiple viral infections were reported at only 10% in a previous study on 40 fresh tissues with primary breast carcinoma.Citation20 Whether multiple viral infections have a role in the outcome of the disease needs further investigations.
Upon investigating the correlation between single and multiple infections in fresh tissues and some clinico-pathological parameters, we found that multiple viral infections were associated with the young age of BC women as reported previouslyCitation16 that may be related to sex hormones.Citation2 Such association needs to be further investigated. High HPV viral load was also associated with young age (≤45y). This may lead to lower expression of breast cancer gene 1 (BRCA1) gene expression and hence early development of BC as previously explained.Citation76,Citation77 Limitations to this study are the relatively small size of samples used. The biological activity of such viruses needs to be further investigated in a larger sample size by exploring the presence of viral transcripts and detection of infectious virions.
Conclusion
Up to our knowledge, this is the first report, in the Arab world, concerning the presence of HPV, EBV, and HMTV and their association with some clinico-pathological characteristics of BC disease. HPV and HMTV have been the most prevalent viruses among studied Egyptian women with BC. The three viruses together (HPV/EBV/HMTV) have existed in about one-third of fresh tissues of studied BC patients. Fresh tissues have been a more favorable specimen than FFPE to detect viral DNAs in BC patients. WBCs could be a more suitable specimen instead of fresh tissue for HMTV detection in BC patients to avoid invasive procedures. Detection of HPV DNA among young women (≤45 years) might be a relevant prognostic marker for the clinical course of BC disease. Further analysis is needed to explore the environmental cause of oncogenic virus infections on a large number of BC patients and controls. In addition, additional analysis is required to investigate the inverse relation between HMTV and HER2 expression and the linkage of HMTV to HPV, EBV, and their biological activities and impact on the outcome of BC disease.
Abbreviations
BHRF1, BamHI-H rightward open reading frame 1; BC, breast cancer; bp, base pair; BRCA1, breast cancer gene 1; Cm, centimeter; E, early; EBER, EBV-encoded small RNA; EBNA, EBV nuclear antigen; EBV, EpsteinBarr virus; Env, envelope; ER, estrogen receptor; FFPE, formalin fixed and paraffin embedded; FG, femtogram; Her2, human epidermal growth factor receptor 2; HMTV, human mammary tumor virus; HPV, human papillomavirus; IDC, invasive ductal carcinoma; ILC, invasive lobular carcinoma; IRP, the institutional review board; L, late; LMP, latent membrane protein; MCF7, Michigan Cancer Foundation-7; MMTV-Ltr, Mouse mammary tumor virus long terminal repeat; N, number; NCI, National Cancer Institute; Ng, nanogram; PBMC, peripheral blood mononuclear cell; PCR, polymerase chain reaction; PR, progesterone receptor; TAE, tris-acetate buffer; UV, ultraviolet; WBCs, white blood cells; μg/mL, microgram per milliliter; μL, microliter; μM, micromolar.
Data Sharing Statement
The datasets used and/or analyzed during the current study available from the corresponding author upon reasonable request.
Ethics Approval and Informed Consent
The study was conducted according to the guidelines of the Declaration of Helsinki, and the research has been approved by the National Cancer Institute-Cairo University, IRB. IRB number 201819039.3, Organization number IORG0003381.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
- Ferlay J, Ervik M, Lam F, et al. Egypt source: globocan 2020. Global Cancer Observatory; 2020. Available from: https://gco.iarc.fr/today/data/factsheets/populations/818-egypt-fact-sheets.pdf. Accessed 129, 2021.
- Alibek K, Kakpenova A, Mussabekova A, Sypabekova M, Karatayeva N. Role of viruses in the development of breast cancer. Infect Agent Cancer. 2013;8(1):1–6. doi:10.1186/1750-9378-8-3223302179
- Lawson JS, Salmons B, Glenn WK. Oncogenic viruses and breast cancer: mouse mammary tumor virus (MMTV), bovine leukemia virus (BLV), human papilloma virus (HPV), and Epstein–Barr virus (EBV). Front Oncol. 2018;8(1):1–18. doi:10.3389/fonc.2018.0000129404275
- Simões PW, Medeiros LR, Pires PDS, et al. Prevalence of human papillomavirus in breast cancer: a systematic review. Int J Gynecol Cancer. 2012;22(3):343–347. doi:10.1097/IGC.0b013e31823c712e22214962
- San TH, Fujisawa M, Fushimi S, et al. Low prevalence of human mammary tumor virus (HMTV) in breast cancer patients from Myanmar. Infect Agent Cancer. 2017;12(1):1–7. doi:10.1186/s13027-017-0130-028053656
- Farahmand M, Monavari SH, Shoja Z, Ghaffari H, Tavakoli M, Tavakoli A. Epstein Barr virus and risk of breast cancer: a systematic review and meta-analysis. Futur Oncol. 2019;15(24):2873–2885. doi:10.2217/fon-2019-0232
- Naushad W, Surriya O, Sadia H. Prevalence of EBV, HPV and MMTV in Pakistani breast cancer patients: a possible etiological role of viruses in breast cancer. Infect Genet Evol. 2017;54:230–237. doi:10.1016/j.meegid.2017.07.01028705719
- Yasmeen A, Bismar TA, Kandouz M, Foulkes WD, Desprez PY, Al Moustafa AE. E6/E7 of HPV type 16 promotes cell invasion and metastasisof human breast cancer cells. Cell Cycle. 2007;6(16):2038–2042. doi:10.4161/cc.6.16.455517721085
- Khodabandehlou N, Mostafaei S, Etemadi A, et al. Human papilloma virus and breast cancer: the role of inflammation and viral expressed proteins. BMC Cancer. 2019;19(1):1–11. doi:10.1186/s12885-019-5286-030606139
- Girianelli VR, Thuler LCS, Silva GAE. Prevalence of HPV infection among women covered by the Family Health Program in the Baixada Fluminense, Rio de Janeiro, Braz. Rev Bras Ginecol e Obs. 2010;32(1):39–46. doi:10.1590/s0100-72032010000100007
- Dossary RA, Alkharsah KR, Kussaibi H. Prevalence of Mouse Mammary Tumor Virus (MMTV)-like sequences in human breast cancer tissues and adjacent normal breast tissues in Saudi Arabia. BMC Cancer. 2018;18(1):1–10. doi:10.1186/s12885-018-4074-629291726
- Naushad W, Ayub S, Sadia H. Significant correlation of MMTV (Mouse mammary tumor virus) LTR gene with hormone receptor status in peripheral blood samples of breast cancer patients from North Pakistan. Int J Biosci. 2017;6655:399–405. doi:10.12692/ijb/10.3.399-405
- Bonnet M, Guinebretiere JM, Kremmer E, et al. Detection of Epstein-Barr virus in invasive breast cancers. J Natl Cancer Inst. 1999;91(16):1376–1381. doi:10.1093/jnci/91.16.137610451442
- Fawzy S, Sallam M, Mohammad N. Detection of Epstein – Barr virus in breast carcinoma in Egyptian women. Clin Biochem. 2008;41:486–492. doi:10.1016/j.clinbiochem.2007.12.01718258188
- Hassab El-Naby NED, Hameda HM, Asmaa MG, Ahmed ESM. Epstein-Barr virus infection and breast invasive ductal carcinoma in Egyptian women: a single center experience. J Egypt Natl Canc Inst. 2017;29(2):77–82. doi:10.1016/j.jnci.2017.02.00228462850
- Glenn W, Heng B, Delprado W, Iacopetta B, Whitaker NJ, Lawson JS. Epstein-Barr virus, human papillomavirus and mouse mammary tumour virus as multiple viruses in breast cancer. PLoS One. 2012;7(11):e48788. doi:10.1371/journal.pone.004878823183846
- Al Moustafa A, Al-Antary N, Al-Antary N, et al. Co-prevalence of Epstein–Barr virus and high-risk human papillomaviruses in Syrian women with breast cancer. Hum Vaccin Immunother. 2016;12(7):1936–1939. doi:10.1080/21645515.2016.113925527082145
- Ahmed RA, Yussif SM. Immunohistochemical detection of human cytomegalovirus, Epstein-Barr virus and human papillomavirus in invasive breast carcinoma in Egyptian women: a tissue microarray study. J Solid Tumors. 2016;6(2):8–16. doi:10.5430/jst.v6n2p8
- El-shinawi M, Mohamed HT, Abdel-fattah HH, Mohamed MM. Inflammatory and non-inflammatory breast cancer: a potential role for detection of multiple viral DNAs in disease progression. Ann Surg Oncol. 2016;23(2):494–502. doi:10.1245/s10434-015-4888-2
- Gad Al Karim AS, Sharaf AEMM, El-Fadeel MA, Al-Shinawi MS, El-Mahdy H. Molecular characterization of viruses involvement in breast cancer patients. Cancer Biol. 2014;4(3):22–34. doi:10.1080/14768320500230185
- Lakhani SR, Ellis IO, Schnitt SJ, Tan PH, Van de Vijvr MJ. WHO Classification of Tumors of the Breast. Pathology and Genetics of Tumors of Breast and Female Genital Organs. 4th ed. IARC Press; 2012.
- Easton DF. Familial risks of breast cancer. Breast Cancer Res. 2002;4(5):179–181. doi:10.1186/bcr44812223120
- Van Der Bij W, Torensma R, Van Son WJ, et al. Rapid immunodiagnosis of active cytomegalovirus infection by monoclonal antibody staining of blood leucocytes. J Med Virol. 1988;25(2):179–188. doi:10.1002/jmv.18902502082839611
- Loutfy SA, Fawzy M, El-Wakil M, Moneer MM. Presence of human herpes virus 6 (HHV6) in pediatric lymphomas: impact on clinical course and association with cytomegalovirus infection. Virol J. 2010;7(1):287. doi:10.1186/1743-422X-7-28720979645
- Manos MM, Ting Y, Wright DK, Lewis AJ, Broker TR, Wolinsky SM. Use of polymerase chain reaction amplification for the detection of genital human papillomaviruses. In: Cancer Cells. 7th ed. Cold Spring Harbor Laboratory Press; 1989:214.
- Hashimoto. Y, Nawata. Y, Kurasawa. K, et al. Investigation of EB VIRUS and cytomegalovirus in rapidly progressive interstetial pneumonitis in polymyositis/dermatomyositis by in situ hyberdization and polymerase chain reaction. Clin Immunol Immunol Pathol. 1995;77(3):298–306. doi:10.1006/clin.1995.1156
- Loutfy SA, Abo-shadi MA, Fawzy M, et al. Epstein-Barr virus and cytomegalovirus infections and their clinical relevance in Egyptian leukemic pediatric patients. Virol J. 2017;2:1–11. doi:10.1186/s12985-017-0715-7
- Wang Y, Holland JF, Bleiweiss IJ, et al. Detection of mammary tumor virus ENV gene-like sequences in human breast cancer. Cancer Res. 1995;55(22):5173–5179.7585568
- Islam S, Dasgupta H, Roychowdhury A, et al. Study of association and molecular analysis of human papillomavirus in breast cancer of Indian patients: clinical and prognostic implication. PLoS One. 2017;12(2):1–17. doi:10.1371/journal.pone.0172760
- Fernandez-Cobo M, Melana SM, Holland JF, Pogo BGT. Transcription profile of a human breast cancer cell line expressing MMTV-like sequences. Infect Agent Cancer. 2006;1(1):1–5. doi:10.1186/1750-9378-1-723509916
- ElSheshtawy NM, Shakweer MM, EL-Ghamrini YM. Is HPV virus associated with human breast cancer? Time to re-examine the postulate. Egyptian Study. Ann Pathol Lab Med. 2017;4(1):A1–A9. doi:10.21276/apalm.2017.1143
- Elashoff JD. nQuery Advisor. Version 7.0 User’s Guide. V edition. Statistical Solutions Ltd.; 2007.
- Habyarimana T, Attaleb M, Baptiste J, Youssef M, Mohammed B, Mzibri E. Detection of human papillomavirus DNA in tumors from Rwandese breast cancer patients. Breast Cancer. 2018;25(2):127–133. doi:10.1007/s12282-018-0831-229350329
- Alshammari AMA, Tarish HR, Aljanabi A. Detection of mouse mammary tumor virus-like sequence (MMTV- like sequence) in the breast cancer of Iraqi women samples by nested PCR. Kufa J Nurs Sci. 2014;4(2):34–143.
- Sigaroodi A, Nadji SA, Naghshvar F, Nategh R, Emami H, Velayati AA. Human papillomavirus is associated with breast cancer in the north part of Iran. Sci World J. 2012;2012:8. doi:10.1100/2012/837191
- Eslamifar A, Ramezani A, Azadmanesh K, Bidari-Zerehpoosh F, Banifazl M, Aghakhani. A. Assessment of the association between human papillomavirus infection and breast carcinoma. Iran J Pathol. 2015;10(1):41–46.26516324
- Bakhtiyrizadeh S, Hosseini SY, Yaghobi R, Sarvari J. Almost complete lack of human cytomegalovirus and human papillomaviruses genome in benign and malignant breast lesions in Shiraz, Southwest of Iran. Asian Pacific J Cancer Prev. 2017;18:3319–3324. doi:10.22034/APJCP.2017.18.12.3319
- Lindel K, Forster A, Altermatt HJ, Greiner R, Gruber G. Breast cancer and human papillomavirus (HPV) infection: no evidence of a viral etiology in a group of Swiss women. Breast. 2007;16(2):172–177. doi:10.1016/j.breast.2006.09.00117088061
- De Cremoux P, Thioux M, Lebigot I, Sigal-Zafrani B, Salmon R, Sastre-Garau X. No evidence of Human papillomavirus DNA sequences in invasive breast carcinoma. Breast Cancer Res Treat. 2008;109(1):55–58. doi:10.1007/s10549-007-9626-417624590
- Hedau S, Kumar U, Hussain S, et al. Breast cancer and human papillomavirus infection: no evidence of HPV etiology of breast cancer in Indian women. BMC Cancer. 2011;11(1):27. doi:10.1186/1471-2407-11-2721247504
- Teo IA, Shaunak S. Polymerase chain reaction in situ: an appraisal of an emerging technique. Histochem J. 1995;27(9):647–659. doi:10.1007/BF02166788557528
- Khan NA, Castillo A, Koriyama C, et al. Human papillomavirus detected in female breast carcinomas in Japan. Br J Cancer. 2008;99(3):408–414. doi:10.1038/sj.bjc.660450218648364
- Chang P, Wang T, Yao Q, Wang L, Chen J. Absence of human papillomavirus in patients with breast cancer in north-west China. Med Oncol. 2012;29:521–525. doi:10.1007/s12032-011-9945-521499926
- Yahia ZA, Adam AAM, Elgizouli M, et al. Epstein Barr virus: a prime candidate of breast cancer aetiology in Sudanese patients. Infect Agent Cancer. 2014;9(1):1–5. doi:10.1186/1750-9378-9-924597902
- Marrão G, Habib M, Paiva A, et al. Epstein-Barr virus infection and clinical outcome in breast cancer patients correlate with immune cell TNF- α/IFN- γ response. BMC Cancer. 2014;14(1):1–11. doi:10.1186/1471-2407-14-66524383403
- Zhang W, Wang M, Wei X, et al. Associations of Epstein-Barr virus DNA in PBMCs and the subtypes with breast cancer risk. J Cancer. 2017;8(15):2944–2949. doi:10.7150/jca.2033028928885
- Zekri AN, Bahnassy AA, Mohamed WS, Hassan ZK. Epstein-Barr virus and breast cancer: epidemiological and molecular study on Egyptian and Iraqi women. J Egypt Natl Canc Inst. 2012;24(3):123–131. doi:10.1016/j.jnci.2012.06.00122929918
- Aboulkassim T, Yasmeen A, Akil N, Batist G, Moustafa AA. Incidence of Epstein – barr virus in Syrian women with breast cancer: a tissue microarray study. Hum Vaccin Immunother. 2015;11(4):951–955. doi:10.1080/21645515.2015.100934225933186
- Wiencke JK. Impact of race/ethnicity on molecular pathways in human cancer. Nat Rev Cancer. 2004;4(1):79–84. doi:10.1038/nrc125714708028
- Glaser SL, Hsu JL, Gulley ML. Epstein-Barr virus and breast cancer: state of the evidence for viral carcinogenesis. Cancer Epidemiol Biomarkers Prev. 2004;13(5):688–697.15159298
- Yasui Y, Potter JD, Stanford JL, et al. Breast cancer risk and “delayed” primary Epstein-Barr virus infection. Cancer Epidemiol Biomarkers Prev. 2001;10(1):9–16.11205495
- Gulley ML. Molecular diagnosis of Epstein-Barr virus-related diseases. J Mol Diagn. 2001;3(1):1–10. doi:10.1016/S1525-1578(10)60642-311227065
- Brink AA, Van Den Brule AJ, Van Diest P, Meijer CJ. Re: detection of Epstein-Barr virus in invasive breast cancers. J Natl Cancer Inst. 1992;84(9):655–656. doi:10.1093/jnci/92.8.655
- Saeedi Z, Hadi F, Hejazi SH, Salahshournia Z. The relationship between EBV virus and breast cancer in Khuzestan Province of Iran. J Appl Biotechnol Rep. 2018;5(1):37–41. doi:10.29252/jabr.01.01.07
- Dowran R, Joharinia N, Safaei A, Bakhtiyarizadeh S, Soleimani AA. No detection of EBV, BKV and JCV in breast cancer tissue samples in Iran. BMC Res Notes. 2019;12(1):1–5. doi:10.1186/s13104-019-4178-330602384
- Sharaf HM, Gomaa MF. Molecular detection of Epstein-Barr virus in breast cancer. Egypt J Hosp Med. 2012;47(1):238–248. doi:10.21608/ejhm.2012.16294
- Morales-sa A, Martı JLE, Mantilla A, Leal YA, Torres J, Fuentes-panana EM. No association between Epstein-Barr virus and mouse mammary tumor virus with breast cancer in Mexican Women. Sci Rep. 2013;3:2970. doi:10.1038/srep0297024131889
- Ribeiro-Silva A. Epstein-Barr virus in breast carcinoma in Argentina. Arch Pathol Lab Med. 2005;129(9):1088. doi:10.5858/2005-129-1088-EVIBCI16119976
- Arbach H, Viglasky V, Lefeu F, et al. Epstein-Barr Virus (EBV) genome and expression in breast cancer tissue: effect of EBV infection of breast cancer cells on resistance to paclitaxel (Taxol). J Virol. 2006;80(2):845–853. doi:10.1128/jvi.80.2.845-853.200616378986
- Herrmann K, Niedobitek G. Lack of evidence for an association of Epstein–Barr virus infection with breast carcinoma. Breast Cancer Res. 2002;5(1):13–17. doi:10.1186/bcr561
- Griffin BE. Epstein-Barr virus (EBV) and human disease: facts, opinions and problems. Rev Mutat Res. 2000;462(2–3):395–405. doi:10.1016/S1383-5742(00)00028-4
- Labrecque LG, Barnes DM, Fentiman IS, Griffin BE. Epstein-Barr virus in epithelial cell tumors: a breast cancer study. Cancer Res. 1995;55(1):39–45.7805038
- Hafez. MM, Hassan. ZK, Kamel MM, Rouby MNE, Zekri ARN. Expression of MMTV-like env gene in Egyptian breast cancer patients. Egypt J Med Microbiol. 2013;22(2):67–72. doi:10.12816/0004943
- Shariatpanahi S, Farahani N, Salehi AR, Salehi R. High prevalence of mouse mammary tumor virus like gene sequences in breast cancer samples of Iranian women. Nucleosides Nucleotides Nucleic Acids. 2017;36(10):621–630. doi:10.1080/15257770.2017.136049829185860
- Lawson. JS, Tran. DD, Carpenter. E, et al. presence of mouse mammary tumour-like virus gene sequences may be associated with morphology of specific human breast cancer. J Clin Pathol. 2006;59(12):1287–1292. doi:10.1136/jcp.2005.03590716698952
- Hachana M, Trimeche M, Ziadi S, et al. Prevalence and characteristics of the MMTV-like associated breast carcinomas in Tunisia. Cancer Lett. 2008;271(2):222–230. doi:10.1016/j.canlet.2008.06.00118639977
- Etkind P, Du J, Khan A, Pillitteri J, Wiernik PH. Mouse mammary tumor virus-like env gene sequences in human breast tumors and in a lymphoma of a breast cancer patient. Clin Cancer Res. 2000;6(4):1273–1278.10778951
- Motamedifar M, Saki M, Ghaderi A. Lack of association of mouse mammary tumor virus-like sequences in Iranian breast cancer patients. Med Princ Pr. 2012;21:244–248. doi:10.1159/000334572
- Tawfik AM, Kassem BEB, Ayoub A, El-baseer MAA. Search for murine mammary tumor virus-like sequences in Egyptian females breast cancer. Am J Med Med Sci. 2015;5(5):223–226. doi:10.5923/j.ajmms.20150505.06
- Lawson JS, Glenn WK. Multiple oncogenic viruses are present in human breast tissues before development of virus associated breast cancer. Infect Agent Cancer. 2017;12(1):1–8. doi:10.1186/s13027-017-0165-228053656
- Park DJ, Southey MC, Giles GG, Hopper JL. No evidence of MMTV-like env sequences in specimens from the Australian Breast Cancer Family Study. Breast Cancer Res Treat. 2011;125:229–235. doi:10.1007/s10549-010-0946-420512660
- Kamal SM, Makki RF, Hammoudi DA. Mouse Mammary Tumor Virus (MMTV) like sequence in breast cancer of Egyptian women. Int J Canc Prev. 2005;2(2):109–115.
- Pao CC, Lin SS, Lin CY, Maa JS, Lai CH, Hsieh TT. Identification of human papillomavirus DNA sequences in peripheral blood mononuclear cells. Am J Clin Pathol. 1991;95(4):540–546. doi:10.1093/ajcp/95.4.5401849701
- Foresta C, Bertoldo A, Garolla A, et al. Human papillomavirus proteins are found in peripheral blood and semen Cd20+ and Cd56+ cells during HPV-16 semen infection. BMC Infect Dis. 2013;13(1):1–10. doi:10.1186/1471-2334-13-59323280237
- Chen. A, Keleher A, Kedda MA, Spurdle AB, McMillan NAJ, Antonsson A. Human papillomavirus DNA detected in peripheral blood samples from healthy Australian male blood donors. J Med Virol. 2009;81(10):1792–1796. doi:10.1002/jmv.2159219697401
- Lawson JS, Glenn WK, Salyakina D, Delprado W, Davidson B, Lawson JS. Human papilloma viruses and breast cancer. Front Oncol. 2015;5(December):1–12. doi:10.3389/fonc.2015.0027725667919
- Polansky H, Schwab H. How latent viruses cause breast cancer: an explanation based on the microcompetition model. Bosn J Basic Med Sci. 2019;19(3):221–226. doi:10.17305/bjbms.2018.395030579323
