Abstract
Background
Since 2015, plasmid-borne mcr-1 has been reported in various bacterial strains in the clinical setting globally. However, the transmission mechanisms of this gene in Salmonella are not well defined. This study aimed to characterize the genomic features of a Salmonella enterica ST34 isolate, which carried a mcr-1, mapped to a carbapenemase and extended spectrum β-lactamase encoding gene located on the IncX4 plasmid.
Methods
Salmonella enterica was recovered from a diarrheal paediatric patient in Shenzhen, China. Antimicrobial susceptibility testing was performed by using the VITEK 2 system. Drug resistance genes were identified using targeted primers and Sanger sequencing. The transferability and genome location of mcr-1 was determined by performing conjugation, S1-PFGE and Southern blot hybridization analysis. WGS was performed by Illumina MiSeq sequencing and was assembled using the A5-Miseq pipeline, and gene annotation was performed using RAST 2.0. The database Centre for Genomic Epidemiology’s website was used to identify resistance genes and sequence types (STs).
Results
We found that the isolate was extensively drug resistant and belonging to ST34, carrying an IncX4 plasmid with mcr-1, blaKPC-2 and blaCTX-M-15. We also noticed that genes blaPAO, fosA, catB, the mutation in oprD and mexT (MexEF-OprN efflux regulator), and exotoxin-encoding genes (exoS, exoY and exoT) were associated with resistance and virulence in the genome. In addition, heavy metal resistance genes as silP and silE were determined.
Conclusion
This study highlights the potential risk of ST34 of Salmonella enterica serotype Typhimurium carrying multiple drug resistance encoding genes in a single IncX4 plasmid.
Introduction
Acute diarrheal diseases are associated with significant mortality and morbidity.Citation1 In the year 2018, the World Health Organization (WHO) reported more than 2 billion people globally that suffered from diarrheal disease.Citation1 Salmonella species are becoming a major global public health concern. These species cause a broad range of clinical conditions, the most common is gastroenteritis, followed by bacteraemia and enteric fever.Citation2 Colistin is commonly used as a last choice of drug to treat infections caused by multi-drug resistant (MDR) or extensively drug-resistant (XDR) bacteria.Citation3 Extended-spectrum beta-lactamase (ESBLs) is prominently encoded by blaCTX-M-15 gene and hydrolyses the penicillin, third-generation cephalosporin. Carbapenemase enzymes encoded by alleles of the blaKPC gene is hydrolysed by the carbapenems antibiotic such as meropenem.Citation4,Citation5 The plasmid-borne colistin resistance mcr-1 was reported in Escherichia coli (E. coli) in the year 2015. Since then, colistin has become the last choice of drug to treat conditions caused by highly resistant pathogen.Citation6 Since 2015, eight additional mcr homologous (mcr-1 to mcr-9) have been reported worldwide, and are chromosome mediated, among them mcr-3.Citation7–9 Salmonella species carrying mcr-1 have been recovered from many specimens including food, animals and clinical in China. The occurrence and distribution of clinical Salmonella enterica serotype Typhimurium (S. Typhimurium) ST34 carrying mcr-1 was found to be low so far.Citation10 Among the most recorded clinical isolates, producing mcr-1 recovered from bloodstream infection and a faecal sample is rare.Citation11 Young children are at high risk of acquiring Salmonella infection due to their underdeveloped immune system.Citation12 The increasing prevalence of MDR Salmonella in pediatric patients poses a serious challenge for treatment due to restricted drug of choice.Citation13 Here, we report a colistin-resistant S. Typhimurium ST34 with IncX4 plasmid carrying mcr-1, blaKPC-2 and blaCTX-M-15, which was isolated from a paediatric patient, suffered suffering from diarrhoea. Antibiogram, horizontal gene transformation and whole-genome sequencing were performed to demonstrate the molecular characteristics of the isolate.
Methods
Bacterial Isolation and Identification
Strain SP-15-127 was isolated from a faecal sample of a patient in 2015, in Shenzhen, China. Species primarily identification was done by using the VITEK 2 compact system (bioMérieux, France), followed by 16S rRNA Sanger sequencing from a commercial company (Sangon Biotech, Shanghai). The full length of 16S rRNA gene was amplified via conventional PCR by using F-5’-GGAACTGAGACACGGTCCAG −3’ and R-5’-CCAGGTAAGGTTCTTCGCGT-3’. PCR reaction volume was 20 µL contained 1µL (30ng) of genomic DNA, 0.4 µL (10 pmol) of each forward and reverse primer, 10 µL of 2X Master Mix and 8.2 µL of nuclease-free water. Thermocycler set for 5 minutes at 95°C for initial denaturation, 35 cycles each 30 sec. 94°C for denaturation, 25 sec at 56°C for annealing and 50 sec at 72°C for extension and final extension at 72°C for 7 minutes, the PCR product was run on 1% agarose along with a DNA ladder. This isolate was collected as a routine Hospital investigation procedure. Only verbal consent was obtained because no personal information was used for research purposes, therefore written consent was not required. All experiments were conducted as per the hospital biosafety regulations act. The polymerase chain reaction (PCR) assay was performed to detect mcr-1, blaKPC-2 and blaCTX-M-15 using specific primers as we previously described.Citation14 Purified PCR products were sequenced (Sanger sequencing method) by Sangon Biotech-Shanghai, China. DNA sequences were analysed by the NCBI-BLAST program (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome).
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility was performed using VITEK 2 compact-60 system with ASTGN09 card and software version 9.01 (bioMérieux) for amikacin, aztreonam, nitrofurantoin, ciprofloxacin, piperacillin, gentamicin, cefepime, ceftriaxone, ceftazidime, tobramycin, imipenem, levofloxacin and sulfamethoxazole/trimethoprim as per manufacture instructions. The E-test method was used to determine the MIC value of meropenem, but for colistin, the broth dilution method was used in accordance with Clinical and Laboratory Standards Institute (CLSI) guidelines. A positive control strain characterized from our laboratory was used, while ATCC25922 was used as a quality control strain.Citation4 Results were interpreted according to CLSI instructions, while colistin resistance was defined according to European Committee on Antimicrobial Susceptibility Testing (EUCAST) clinical breakpoints.Citation15,Citation16
Conjugation, CeuI-PFGE and Southern Hybridization
Conjugation was performed using the isolate SP-15-127 as a donor cell while streptomycin-resistant Escherichia coli strain C600 (E. coli C600) was used as the recipient. The broth medium for both isolates were mixed and incubated for meeting at 37° C for 24 hrs as described previously.Citation17 The conjugants were screened on Muller Hinton agar plates containing colistin (4 μg/mL) and streptomycin (2000 μg/mL) for 18 hrs. Thereafter, conjugants were selected for antibiotic susceptibility to determine the phenotypic expression of transferred genes followed by PCR assay and sequencing (Sanger sequencing). The plasmid and (or) chromosomal locations of mcr-1 were determined by S1-PFGE, followed by southern hybridizations. The genomic DNA of the isolate SP-15-127 was digested with S1 endonuclease (Takara Biotech).Citation18 The digested DNA fragments were separated by CHEFDR III BioRad system with a run time of 12 hrs and switch time of 5–40 Seconds, and lambda ladder was used as a marker. Southern hybridizations of plasmid DNA were performed with a digoxigenin-labelled mcr-1 probe, according to the manufacturer’s instructions (Roche Diagnostics, Germany).
Whole-Genome Sequencing and Bioinformatic Analysis
The isolated Salmonella enterica serotype Typhimurium strain (SP-15-127) was subjected to whole-genome extraction using the Qiagen Blood & Tissue kit (Qiagen, Hilden, Germany, Lot No. 121223). DNA quantity and quality were analysed using gel electrophoresis and the BioDrop DUO UV/VIS spectrophotometer device (BioDrop England). The DNA library was prepared with 400bp paired-end fragment and sequencing was performed using an Illumina HiSeq 2000 platform. The mixed assembly of Illumina was assembled into contigs using SPAdes version 3.11.1 and the quality of the assemblies was evaluated using the software QUAST. Plasmid incompatibility, multi-locus sequences typing (MLST), antimicrobial resistance genes (AMR) and virulence genes were identified using the Centre for Genomic Epidemiology (CGE) platform (http://www.genomicepidemiology.org/services/).Citation19
Results
Isolate SP-15-127 Characteristics
Isolate SP-15-127 was conferred as S. Typhimurium which was recovered from a faecal sample of a 6-year male child diagnosed with gastroenteric and bacteraemia. The PCR assay demonstrated that SP-15-127 harbouring colistin resistance mcr-1, carbapenemase encoding blaKPC-2 and extended β-lactamase encoding blaCTX-M-15 genes which hydrolysed the ceftazidime and ceftriaxone. Antibiogram results indicated that isolate SP-15-127 was resistant to most antibiotics including colistin, aztreonam, nitrofurantoin, ciprofloxacin, piperacillin, cefepime, ceftriaxone, ceftazidime, levofloxacin, and imipenem but sensitive to the tobramycin, tigecycline ().
Table 1 Antimicrobial Susceptibility of Salmonella Sp-15-127, Transconjugant’s and E. Coli C600 (Recipient)
Transferability and Location of Genes
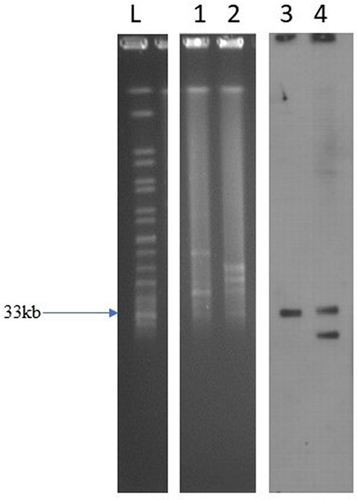
Performing the conjugation experiments, we found that isolate was able to transfer their colistin resistance phenotype to E. coli C600. The frequency of conjugation for the mcr-1 gene was 5.6% in isolates. S1-PFGE and Southern hybridization indicated that mcr-1 gene was located on a 40kb plasmid belonging to IncX4 (). The resistant phenotype showed that conjugants were resistant to aztreonam, piperacillin, cefepime, ceftriaxone, ceftazidime, and imipenem (). The PCR product sequencing revealed that both blaKPC-2 and blaCTX-M-15 was also located on the same plasmid.
Figure 1 S1-PFGE pattern for SP-15-127 strain and Southern blot analysis of mcr-1 genes.
Genome Characterization
MLST results revealed that S. Typhimurium belongs to the ST34 group. The isolate of a single plasmid belonging to the Incx4 was confirmed using Plasmid Finder. The genetic context of the MCR-1, CTX-M-15 and KPC-2 in the plasmid was represented in (Supplementary Material 1). Our genomic characterization data reports the plasmid-encoded resistance genes, such as colistin aminoglycoside [aadA1 and aac(3)-IVa], trimethoprim (dfrA12), sulphonamide (sul1, sul2 and sul3), phenicol (floR and cmlA1) and fosfomycin (fosA3). We also found evidence of IncX3 plasmid harbouring the blaTEM-1 (cephalosporin resistance), floR (florfenicol resistance), tet(A) (tigecycline resistance) and blaSHV-12 (carbapenem resistance) genes. Most of the resistance genes are located on the plasmids, indicating transferability and transmission of the corresponding resistance genes to other bacteria. Copper resistance gene operons (cus operon and cop operon) and silver resistance genes (silP and silE) were reported, which was found to be responsible for the heavy metal resistance in this isolate.
Discussion
The prevalence of colistin-resistant bacteria is increasing attention, therefore considered as a threat to global health.Citation20 Colistin is the last choice of drug to treat conditions caused by carbapenemase-producing bacteria.Citation21 Nevertheless, we found the emergence of the plasmid-borne colistin resistance gene mcr-1 co-existence and co-transmission with extended beta-lactamase encoding genes as well as carbapenemase encoding genes such as blaKPC, and blaCTX-M, which are located on transferable element plasmid.Citation22,Citation23 The prevalence of mcr-1 harbouring S. Typhimurium is highly in animal husbandry but still low in humans, specifically in children.Citation24,Citation25 The K. pneumoniae highest mcr-1-positive rate is about 10%, which is quite higher than salmonella in China.Citation26 Furthermore, the co-existence of mcr-1 (blaKPC and blaCTX-M) in one S. Typhimurium strain is not yet reported in China. In this study, only one isolate among nearly thousands of clinical S. Typhimurium strains was confirmed as a multidrug-resistant superbug that harboured these three important resistance genes. Unfortunately, the mcr-1, blaKPC-1 and blaCTX-M-15 genes were located on a single plasmid, which has a high possibility of their co-transfer. The multidrug-resistant strain SP-15-127 belongs to ST34 based on the S. Typhimurium MLST scheme. Previous studies on S. Typhimurium ST34 were associated with copper resistance and toxin production in the gut.Citation27,Citation28 In addition, ST34 strains were found to be a dominant host for the mcr-1-IncX4 plasmid, which possessed a highly conserved sequence and plasmid structure. Moreover, the carbapenemase gene blaNDM, and blaCTX-M has been reported occasionally, but blaKPC emerged rarely in the ST34 strain.Citation29 Plasmids play a key role in harbouring and transferring resistance genes, especially in Enterobacteriaceae.Citation4 We harvested single plasmid IncX4 from SP-15-127 strain, which encode resistance genes including mcr-1, blaKPC-2, and blaCTX-M-15. Luo et al have reported the presence of S. Typhimurium ST34 carrying mcr-1 in a paediatric patient with bloodstream infection, and the genomic location was pHNSHP45-2-like IncHI2 plasmid.Citation9 In this study, the mcr-1 harbouring plasmid IncX4 was highly distributed in E. coli, suggesting that the transmission of plasmid may occur from E. coli to Salmonella. The mcr-1 location on IncX4 plasmid is considered a major reservoir and is highly disseminated in China, both in humans and the environment.Citation30 Our reported plasmid has a high transfer rate (5.6%) which was similar to the one reported by Jian et al. In their study, they screened IncX4 plasmids among 2470 isolates of Enterobacteriaceae and determined the transfer rate.Citation31 One of the potential limitations of this study is the whole plasmid sequence.
Conclusion
Here, we report the occurrence of ST34 of Salmonella enterica serotype Typhimurium carrying multiple drug resistance encoding genes co-harbouring mcr-1, blaKPC-2 and blaCTX-M-15 in IncX4 plasmid. These resistant determinants present on the mobile element of plasmid, potentially, transmit genes via horizontal gene transfer and were responsible for the emergence of the superbug. Our findings also suggest the need for epidemiological surveillance and monitoring of Salmonella superbug transmission emergence.
Data Sharing Statement
All data files mentioned in this manuscript are available.
Ethics Approval and Consent to Participate
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Ethics Committee, Shenzhen Children’s Hospital, Reference number: 2018 (013) on dated 2018/09/03.
Informed Consent Statement
Due to the retrospective nature of the study, the Ethics Committee of Shenzhen Children’s Hospital, Shenzhen determined that patients’ consent was not required. The clinical isolate samples used in this research were part of the routine hospital laboratory procedure No personal patient’s information was used, data were kept confidentially and in compliance with the Declaration of Helsinki.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and also agree to be accountable for all aspects of the work.
Disclosure
The authors declare no conflicts of interest in this work.
Acknowledgments
We would like to thank Prof. Ma Lian and Dr. Chen Xiaowen from the pediatric research institute, Shenzhen Children’s Hospital for their constant support of this project and Dr. Sunil Kumar Shahu from BGI, Shenzhen, for genome assembly.
Additional information
Funding
References
- Besser JM. Salmonella epidemiology: a whirlwind of change. Food Microbiol. 2018;71:55–59. doi:10.1016/j.fm.2017.08.018
- Eng S-K, Pusparajah P, Ab Mutalib N-S, Ser H-L, Chan K-G, Lee L. Salmonella: a review on pathogenesis, epidemiology and antibiotic resistance. Front Life Sci. 2015;8(3):284–293. doi:10.1080/21553769.2015.1051243
- World Health Organization. Critically important antimicrobials for human medicine, 5th revision; 2017. Available from: https://www.who.int/foodsafety/publications/antimicrobials-fifth/en/. Accessed February 22, 2022.
- Patil S, Chen X, Lian M, Wen F. Phenotypic and genotypic characterization of multi-drug-resistant Escherichia coli isolates harboring blaCTX-M group extended-spectrum β-lactamases recovered from pediatric patients in Shenzhen, southern China. Infect Drug Resist. 2019;12:1325–1332. doi:10.2147/IDR.S199861
- Patil S, Chen X, Wen F. Exploring the phenotype and genotype of multi-drug resistant Klebsiella pneumoniae harbouring blaCTX-M group extended-spectrum β-lactamases recovered from paediatric clinical cases in Shenzhen, China. Ann Clin Microbiol Antimicrob. 2019;18:32. doi:10.1186/s12941-019-0331-z
- Liu Y, Wang Y, Walsh T, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi:10.1016/S1473-3099(15)00424-7
- Carattoli A, Villa L, Feudi C, et al. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill. 2017;22(31):30589. doi:10.2807/1560-7917.ES.2017.22.31.30589
- Borowiak M, Fischer J, Hammerl J, Hendriksen R, Szabo I, Malorny B. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi-B. J Antimicrob Chemother. 2017;72(12):3317–3324. doi:10.1093/jac/dkx327
- Carroll M, Gaballa A, Guldimann C, Sullivan G, Henderson O, Wiedmann M. Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype typhimurium isolate. mBio. 2019;10(3):e00853–e00919.
- Li X-P, Fang L-X, Song J-Q, et al. Clonal spread of mcr-1 in PMQR-carrying ST34 Salmonella isolates from animals in China. Sci Rep. 2016;6(1):38511. doi:10.1038/srep38511
- Luo Q, Wan F, Yu X, et al. MDR Salmonella enterica serovar Typhimurium ST34 carrying mcr-1 isolated from cases of bloodstream and intestinal infection in children in China. J Antimicrob Chemother. 2020;75(1):92–95. doi:10.1093/jac/dkz415
- Woh Y, Yeung P, Nelson E, Goggins WBIII. Risk factors of non-typhoidal Salmonella gastroenteritis in hospitalised young children: a case–control study. BMJ Paediatric. 2021;5(1):e000898. doi:10.1136/bmjpo-2020-000898
- Chang Y-J, Chen C, Feng Y, et al. Highly antimicrobial-resistant Nontyphoidal Salmonella from retail meats and clinical impact in children, Taiwan. Pediatr Neonatol. 2020;61(4):432–438. doi:10.1016/j.pedneo.2020.03.017
- Sandip P, Jiang M, Wen F. Molecular characterization of co-existence of MCR-1 and NDM-1 in extended-spectrum β-Lactamase-producing Escherichia coli ST648 isolated from a colonized patient in China. Jundishapur J Microbiol. 2019;12(7):e91272.
- M100-S25 performance standards for antimicrobial susceptibility testing; Twenty-fifth informational supplement; 2015.
- The European Committee on Antimicrobial Susceptibility Testing and Clinical and Laboratory Standards Institute Recommendations for MIC determination of colistin (polymyxin E) as recommended by the joint CLSI-EUCAST Polymyxin Breakpoints Working Group; 2016.
- Long H, Feng Y, Ma K, Liu L, McNally A, Zong Z. The co-transfer of plasmid-borne colistin-resistant genes mcr-1 and mcr-3.5, the carbapenemase gene blaNDM-5 and the 16S methylase gene rmtB from Escherichia coli. Sci Rep. 2019;9(1):696. doi:10.1038/s41598-018-37125-1
- Luo Q, Yu W, Zhou K, et al. Molecular epidemiology and colistin-resistant mechanism of mcr-positive and mcr-negative clinical isolated Escherichia coli. Front Microbiol. 2017;8:2262. doi:10.3389/fmicb.2017.02262
- Larsen V, Cosentino S, Rasmussen S, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. 2012;50(4):1355–1361. doi:10.1128/JCM.06094-11
- Grégoire N, Aranzana-Climent V, Magréault S, Marchand S, Couet W. Clinical pharmacokinetics and pharmacodynamics of colistin. Clin Pharmacokinet. 2017;56(12):1441–1460. doi:10.1007/s40262-017-0561-1
- Andrade F, Silva D, Rodrigues A, Pina-Vaz C. Colistin update on its mechanism of action and resistance, present and future challenges. Microorganisms. 2020;8(11):1716. doi:10.3390/microorganisms8111716
- Zhao D, Zhou Z, Hua X, et al. Co-existence of mcr-1, blaKPC-2 and two copies of fosA3 in a clinical Escherichia coli strain isolated from urine. Infect. Genet. Evol. 2018;60:77–79. doi:10.1016/j.meegid.2018.02.025
- Cao X, Zhong Q, Guo Y, et al. Emergence of the coexistence of mcr-1, blaNDM-5, and blaCTX-M-55 in Klebsiella pneumoniae ST485 clinical isolates in China. Infect Drug Resist. 2021;14:3449–3458. doi:10.2147/IDR.S311808
- Ana B, Grazielle R, Mylenna P, Rafaela F, Carlos A, Pedro P. Global distribution of plasmid-mediated colistin resistance mcr gene in Salmonella: a systematic review. J Appl Microbiol. 2021;00:1–18.
- Liu G, Qian H, Lv J, et al. Emergence of mcr-1-harboring Salmonella enterica serovar sinstorf type ST155 isolated from patients with diarrhoea in Jiangsu, China. Front Microbiol. 2021;12:723697. doi:10.3389/fmicb.2021.723697
- Zhang R, Liu L, Zhou H, et al. Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine. 2017;19:98–106. doi:10.1016/j.ebiom.2017.04.032
- Branchu P, Charity O, Bawn M, et al. SGI-4 in monophasic salmonella typhimurium ST34 is a novel ICE that enhances resistance to copper. Front Microbiol. 2019;10:1118. doi:10.3389/fmicb.2019.01118
- Sana G, Flaugnatti N, Lugo A, et al. Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. PNAS. 2016;113:e5044–e5051. doi:10.1073/pnas.1608858113
- Kai Z, Haojie G, Jingjing H, et al. Salmonella Typhimurium ST34 isolate was more resistant than the ST19 isolate in China, 2007 − 2019. Foodborne Pathog Dis. 2021;Ahead of Print:45.
- Sun J, Fang L-X, Wu Z, et al. Genetic analysis of the IncX4 plasmids: implications for a unique pattern in the mcr-1 acquisition. Sci Rep. 2017;7(1):424. doi:10.1038/s41598-017-00095-x
- Wang R, van Dorp L, Shaw P, et al. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat Commun. 2018;9(1):1179. doi:10.1038/s41467-018-03205-z
