Abstract
Purpose
Helminths and their products can regulate immune response and offer new strategies to control and alleviate inflammation, including asthma. We previously found that a peptide named as SJMHE1 from Schistosoma japonicum can suppress asthma in mice. This study mainly investigated the molecular mechanism of SJMHE1 in inhibiting asthma inflammation.
Methods
SJMHE1 was administered to mice with OVA-induced asthma via subcutaneous injection, and its effects were detected by testing the airway inflammation of mice. The Th cell distribution was analyzed by flow cytometry. Th-related transcription factor and cytokine expression in the lungs of mice were analyzed using quantitative real-time PCR (qRT-PCR). The expression of miR-155 and levels of phosphorylated STAT3 and STAT5 were also determined after SJMHE1 treatment in mice by qRT-PCR and Western blot analysis. The in vitro mouse CD4+ T cells were transfected with lentivirus containing overexpressed or inhibited miR-155, and the proportion of Th17, Treg cells, CD4+p-STAT3+, and CD4+p-STAT5+ cells were analyzed by flow cytometry.
Results
SJMHE1 ameliorated the airway inflammation of asthmatic mice, upregulated the proportion of Th1 and Treg cells, and the expression of Th1 and Treg-related transcription factor and cytokines. Simultaneously, SJMHE1 treatment reduced the percentage of Th2 and Th17 cells and the expression of Th2 and Th17-related transcription factor and cytokines. SJMHE1 treatment decreased the expression of miR-155 and p-STAT3 but increased p-STAT5 expression. In vitro, the percentage of Th17 and CD4+p-STAT3+ cells increased in CD4+ T cells transfected over-expression of miR-155, but SJMHE1 inhibited the miR-155-mediated increase of Th17 cells. Furthermore, SJMHE1 increased the proportion of Treg and CD4+p-STAT5+ cells after transfected over-expression or inhibition of miR-155.
Conclusion
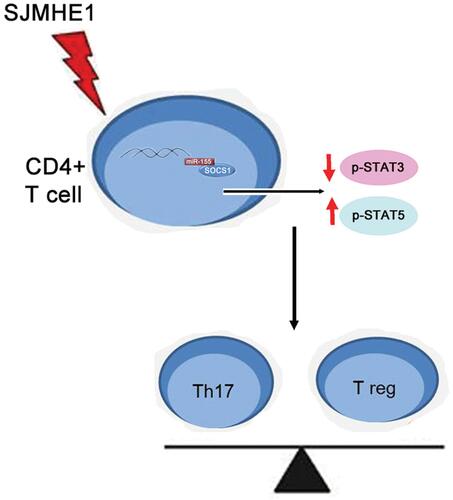
SJMHE1 regulated the balance of Th17 and Treg cells by modulating the activation of STAT3 and STAT5 via miR-155 in asthma. SJMHE1 might be a promising treatment for asthma.
Introduction
Asthma is a heterogeneous disease characterized by chronic airway inflammation and caused by various immune cells and inflammatory factors.Citation1 Although the exact molecular mechanism of asthma remains unknown, CD4+ T cells, especially the imbalance of Th1/Th2 and Th17/regulatory T cells (Tregs), participate in asthma pathogenesis.Citation2,Citation3 Although inhaled corticosteroids are effective for the treatment of asthmatic patients, many side effects and resistance are caused by corticosteroids in patients,Citation4,Citation5 prompting us to seek new therapeutic targets in asthma.
Helminth infection and helminth-derived products reduce allergic, autoimmune, and inflammatory reactions, including asthma.Citation6–Citation8 Helminths have developed several mechanisms to modulate the immune response of hosts, thereby inhibiting the occurrence of asthma.Citation9,Citation10 Helminth infection or helminth-derived molecules induce regulatory T cells (Tregs),Citation11–Citation13 IL-10-producing regulatory B cells,Citation14,Citation15 dendritic cells,Citation16 and alternatively activated macrophagesCitation17 to prevent allergic airway inflammation in asthmatic mice. In comparison with helminth infection or most macromolecules derived from parasites, we have identified a small-molecule peptide from Schistosoma japonicum (S. japonicum) and it named as SJMHE1; it could induce CD4+CD25+ Treg cellsCitation18 and suppress the delayed-type hypersensitivity in mice.Citation19 Furthermore, SJMHE1 can inhibit the inflammatory response in collagen-induced arthritis (CIA)Citation20 and asthmaCitation21 in mice. Asthma was induced by OVA in mice, they were treated by SJMHE1 at the time of OVA administration (day 0), and SJMHE1 suppressed airway inflammation in asthma by downregulating Th2 cells and upregulating Th1 and Treg cells in asthmatic mice.Citation21 However, the molecular mechanism of the downregulation of inflammation in asthma caused by SJMHE1 is not well understood.
Recent studies reveal that non-coding RNAs (ncRNA), especially microRNAs (miRNAs), participate in the regulation of inflammatory response in asthma.Citation22,Citation23 Asthmatic patients up-regulates miR-498, miR-187, and miR-143 and down-regulates miR-18a, miR-126, miR-155, and miR-224 in nasal biopsies.Citation24 The upregulation of miR-155, miR-21, and miR-18a is correlated with Th2 cytokines in bronchoalveolar lavage fluid of asthmatic mice.Citation25 Among these miRNAs involved in the pathogenesis of asthma, miR-155 is an important immune modulator and is upregulated in various activated immune cells.Citation26 Furthermore, miR-155 regulates Th2, Th17, and Treg differentiationCitation27–Citation29 and participates in the pathogenesis of various diseases, including asthma.Citation26 Malmhäll and colleague reported that miR-155 knockout (KO) mice decrease eosinophilic inflammation and mucus hypersecretion and reduce the levels of Th2 cells and Th2-related cytokines, suggesting that miR-155 is essential for Th2 response in asthma.Citation30 miR-155 also regulates Th1/Th2 balance. The overexpression of miR-155 in CD4+ T cells promotes Th1 differentiation, whereas the inhibition of miR-155 induces Th2 response.Citation27 Furthermore, miR-155 is involved in the regulation of Th17/Treg balance in various inflammations.Citation29,Citation31,Citation32 Whether miR-155 is involved in the regulation of Th cells in asthmatic mice by SJMHE1 needs further analysis.
In the present study, we further investigated the role of SJMHE1 in the development of asthma and treated mice with SJMHE1 at days 14 and 21 beginning at the third OVA sensitization. We demonstrate that the SJMHE1 treatment still suppresses the inflammation of the airway and regulates Th cell distribution, the expression of key transcription factors, and cytokines for Th cell differentiation in the lungs of asthmatic mice. Furthermore, SJMHE1 treatment downregulated miR-155, and this condition may suppress airway inflammation by regulating Th17/Treg balance in the asthmatic mice.
Materials and Methods
Mice
Male BALB/C mice aged 6–8 weeks were obtained from Comparative Medicine Centre of Yangzhou University (Yangzhou, China) and bred with specific pathogen-free condition at the Animal Experimental Centre of Jiangsu University. Mouse handling and experimentation were conducted in strict compliance with the Regulations for Administration of Affairs Concerning Experimental Animals of Jiangsu University. All protocols were approved by the Jiangsu University Institutional Animal Care and Use Committee (Permit Number: JSU 18–012).
Peptides
The synthesis and purification of SJMHE1 peptide from SjHSP60 437‐460 (VPGGGTALLRCIPVLDTLSTKNED) are entrusted to Chinapeptides (Shanghai, China). Polymyxin B‐agarose was used to remove possible LPS contamination as described previously.
Induction of Experimental Asthma and SJMHE1 Treatment
Asthma was induced in mice as described previously.Citation21 In brief, mice were randomly divided into four groups, namely, PBS, OVA, OVA/PBS, and OVA/SJMHE1 group. On days 0, 7, and 14, each mouse in PBS group was immunized by intraperitoneal injection of 200 µL of PBS, while the mice in the other groups were immunized by 50 µg OVA (Sigma-Aldrich, Steinheim, Germany) and 2 mg of 10% aluminum hydroxide gel in PBS. On days 14 and 21, mice in OVA/PBS and OVA/SJMHE1 groups were separately treated with PBS and SJMHE1 (10 µg) emulsified with incomplete Freund’s adjuvant (Sigma, Poole, UK) as described previously.Citation21 On days 21–28, the mice were challenged in the form of atomization with OVA (2%) or PBS as previously described.Citation21 All the mice were sacrificed on the day 29 to evaluate airway inflammation and immune response.
Histopathologic Analysis
The left lung of mice was soaked in 10% formalin for paraffin embedding. The lung tissue was then stained with hematoxylin and eosin (H&E) and periodic acid-Schiff (PAS) to determine the histopathologic analysis.
Cell Analysis in Bronchoalveolar Lavage Fluid (BALF)
The left main bronchus was ligated, and the right lung of each mouse was washed twice with 0.5 mL of sterile phosphate buffer solution (PBS) to collect BALF. BALF was centrifuged, and the supernatant was removed. Cell precipitation was resuspended with 1 mL of PBS, and the total number of inflammatory cells was counted using a hemocytometer. Wright and Giemsa staining was used to count the eosinophils in BALF as described previously.Citation21
Anti‐OVA‐specific IgE Detection in Serum
Anti‐OVA‐specific IgE was detected using an enzyme-linked immunosorbent assay (ELISA) as described previously.Citation21 In brief, the plate was coated with 100 µL of OVA (100 µg/mL) per well and blocked with 5% skim milk. After washing, goat anti‐mouse IgE (Abcam, 1:250 diluted) and HRP‐conjugated rabbit anti‐goat secondary IgG (Multisciences, 1:5000 diluted) were used for detection.
Flow Cytometry
Single-cell suspensions of the lungs were prepared and analyzed for Th1, Th2, TH17, and Treg as previously described.Citation21 For Th1, Th2, and Th17 cell subset analysis, lung cell suspensions were stimulated with PMA/Ionomycin mixture (Multisciences, Hangzhou, China) and BFA/Monensin Mixture (MultiSciences) for 5 h, and then stained with PerCP anti-CD3 mAbs (eBioscience, San Diego, CA, USA) and FITC-anti-CD4 (eBioscience). After fixation and permeabilization with Cytofix/Cytoperm (BD Biosciences, San Jose, CA, USA), the cells were stained with APC-anti-IFN-γ (eBioscience), APC-anti-IL-4 (eBioscience), and PE-anti-IL-17A (BioLegend, California, USA). Mouse regulatory T cell staining kit (eBioscience) was used to determine Treg cells as previously described.Citation21
For the detection of CD4+p-STAT3+ and CD4+p-STAT5+ cells, the CD4+T cells were separately stimulated with 100 ng/mL IL-6 (Pepro Tech, Cranbury, NJ, USA) and 100 ng/mL IL-2 (Pepro Tech, Cranbury, NJ, USA) at 37°C for 30 mins following the manufacturer’s protocol. Then, the cells were collected and stained with BV421-anti-CD4 (eBioscience). After fixation and permeabilization with Cytofix/Cytoperm (BD Biosciences), the cells were stained with PE-anti-p-STAT3 (BD Biosciences) and PE-anti-p-STAT5 (BD Biosciences), respectively.
For the detection of CD4+SOCS1+ cells, the CD4+T cells were stained with BV421-anti-CD4 (eBioscience), and then fixed and permeabilized with Cytofix/Cytoperm (BD Biosciences). Then, the cells were collected and added with anti-SOCS1 (Abcam, Cambridge, UK) antibody for 30 mins. After washing, the cells were stained with Alexa Fluor 647 Donkey anti-rabbit IgG (Biolegend). All samples were tested using BD FACSCanto flow cytometer (BD Biosciences) and analyzed with Flowjo software (Tree Star, Ashland, OR, USA).
Quantitative Real-Time PCR (qRT-PCR)
Total RNA extracted from the lungs of mice and CD4+ T cells was reverse-transcribed into cDNA by using the Prime Script 1st Strand cDNA synthesis kit (Takara) and All-in-oneTM miRNA First-Strand cDNA synthesis kit 2.0. All-in-One™ qPCR primer for GAPDH (MQP027158), IFN-γ (MQP027401), IL-4 (MQP032451), IL-5 (MQP029462), IL-13 (MQP092962), IL-17A (MQP029457), IL-10 (MQP029453), IL-35 (MQP027412), TGF-β (MQP030343), T-bet (MQP034192), GATA3 (MQP027165), FoxP3 (MQP067272), ROR-γt (MQP054636), miR-155-5p (MmiRQP0890), and U6 (MmiRQP9002) were obtained from GeneCopoeia (GeneCopoeia, Germantown, MD, USA). The PCR amplification of miR-155-5p was conducted using the All-in-oneTM miRNA qPCR kit (GeneCopoeia, Germantown, MD, USA), and the other primers were analyzed via RT-PCR with ChamQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China) according to the manufacturer’s protocol.
Western Blot Analysis
Protein samples were extracted from mouse lungs for Western blot analysis as previously described.Citation21 The rabbit monoclonal antibodies of p-Stat3 (Tyr705; Cell Signaling Technology; 1:1000 dilution), p-Stat5 (Tyr694; Cell Signaling Technology; 1:1000 dilution), SOCS1 (Cell Signaling Technology; 1:1000 dilution), and GAPDH (Cell Signaling Technology; 1:1000 dilution) were used as the primary antibodies. HRP-conjugated anti-rabbit IgG (AB Clonal, Wuhan, China) was used as the second antibody. The chemiluminescent detection and image analysis were carried out as previously described.Citation21
CD4+ T Cell Isolation and Activation
CD4+ T cells were separated from the splenocyte suspension of naïve mice by using the EasySep™ Mouse CD4+ T cell isolation kit (Stem Cell Technologies, Vancouver, Canada) following the manufacturer’s protocol. The purity of CD4+ T cells was >95% as determined by flow cytometry. CD4+ T cells (2×106/mL) were cultured in the presence or absence of MACSiBead Particles (T cell activation/expansion kit, Miltenyi Biotec, Bergisch Gladbach, Germany) for 24 h. MACSiBead particles are loaded with mouse CD2, CD3, and CD28 antibodies.
CD4+ T Cell Transfection by Lentivirus
Upregulated (overexpression) and knocked down miR-155 (inhibition) in the GV369 and GV280 lentivirus vector (Genechem, Shanghai, China) were transfected with CD4+ T according to the manufacturer’s instructions. In brief, according to the MOI and the titer of lentivirus, 150 µL of GV369, 56 µL of GV280 lentivirus vector, and 20 µL of 250× Histrans G P (Genechem) were added into CD4+ T cells (5 × 105 cells/well) containing 500 µL of PRMI 1640 in a 24-well plate. After 24 h, the medium was replaced by 1 mL of RPMI 1640 containing 10% FBS. The cells were then stimulated by 5 µL of loaded anti-biotin MACSiBead particles (Miltenyi Biotec) for 96 h. At the same time, certain wells were added with 1 µg/mL SJMHE1. Then, the percentages of Th17 and Treg cells, and p-STAT3, p-STAT5, and SOCS1 expression levels in CD4+ T cells were analyzed by flow cytometry.
Statistical Analyses
GraphPad Prism 5.01 (GraphPad Software, 2007, La Jolla, CA, USA) was used for statistical analyses. Data were expressed as mean ± standard error of the mean. One-way ANOVA with Tukey Kramer was used to compare the differences among the groups. Spearman correlation was used for the correlation analysis, and P-value < 0.05 was considered statistically significant.
Results
SJMHE1 Treatment Ameliorated Ongoing Asthma Inflammation in Allergic Mice
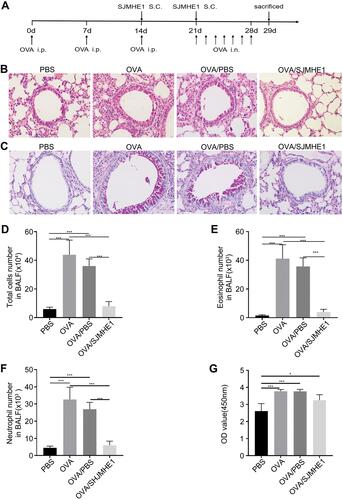
We further investigated whether SJMHE1 could treat ongoing asthma in mice.Citation21 SJMHE1 treatment began with the third OVA sensitization, and the treatment regimen is illustrated in . Consistent with previous results,Citation21 the infiltration of inflammatory cell in the peribronchial and perivascular regions, mucus hypersecretion, structural changes in lungs, such as goblet cell hyperplasia, and increased airway smooth muscle mass, were displayed in OVA- or OVA/PBS-treated mice ( and ). By contrast, inflammatory cell infiltration, mucus secretion, and structural changes in the airway were ameliorated in OVA/SJMHE1 groups. Similarly, SJMHE1 treatment reduced the total cell, eosinophilia, and neutrophil numbers in the BALF () but did not affect the IgE levels relative to the OVA or OVA/PBS group (). These results suggest that SJMHE1 treatment can ameliorate ongoing asthma in mice.
Figure 1 SJMHE1 treatment alleviates ongoing airway inflammation in allergic mice. (A) Experimental scheme. BALB/c mice were sensitized with OVA on days 0, 7, and 14 and challenged with OVA daily from day 21 to day 28. Mice were injected with SJMHE1 emulsified in IFA on days 14 and 21. The mice were killed on day 29. (B) Lung sections from mice were stained by hematoxylin and eosin (H&E), and (C) periodic acid-Schiff reagent (PAS) at 20× magnification. Images represent three independent experiments (n = 6 mice per group). (D) Total cells, (E) eosinophil number, and (F) neutrophil number in BALF. Eosinophil and neutrophil in BALF cells were determined by Wright and Giemsa staining. (G) OVA-specific IgE antibody levels in sera of mice. Results are presented as mean±SEM (n=18) from three-independent experiments. *P < 0.05, ***P < 0.001.
SJMHE1 Treatment Modulates Th Cell Distribution and Transcription Factor and Cytokine Expression in Lungs of Asthmatic Mice
The lungs serve as the inflammatory site in asthmatic mice, and CD4+ T cells are at the forefront of airway inflammation in asthma.Citation2 We investigated the effect of SJMHE1 treatment on the Th cell distribution in the lungs of asthmatic mice. Consistent with previous results in the splenocytes of allergic mice,Citation21 the proportion of Th1 and Treg cells increased in SJMHE1-treated mice compared with OVA or OVA/PBS-treated mice (). Similarly, the percentage of Th2 and Th17 cells decreased in SJMHE1-treated mice compared with OVA or OVA/PBS-treated mice (). T-bet, GATA3, RORγt, and Foxp3 are the dominant transcription factors of Th1, Th2, Th17, and Treg cells, respectively. Consistent with Th cell distribution, SJMHE1 treatment displayed upregulation of T-bet and Foxp3 mRNA expression and downregulation of GATA3 and RORγt mRNA expression (). Consistent with the regulation of Th cells by SJMHE1, SJMHE1 treatment upregulated the expression of IFN-γ, IL-10, TGF-β, and IL-35 mRNA and downregulated the expression of IL-4, IL-5, IL-13, and IL-17 mRNA in the lungs of asthmatic mice (). Therefore, SJMHE1 ameliorated ongoing asthma, which was associated with the regulation of Th differentiation and cytokine expression in the lungs of asthmatic mice.
Figure 2 SJMHE1 treatment regulates Th cell distribution in the lungs of allergic mice. On day 29, the mice were killed, and the lungs of mice were tested for Th1/Th2/Th17/Treg subsets by flow cytometry. (A) CD4+IFN-γ+ Th1 cells, (B) CD4+IL-4+ Th2 cells, (C) CD4+IL-17+ Th17 cells, and (D) CD4+CD25+Foxp3+ Tregs in each group are shown. Data are representative of the experiments. (E) The proportion of CD4+IFN-γ+ Th1 cells, (F) CD4+IL-4+ Th2 cells, (G) CD4+IL-17+ Th17 cells, and (H) CD4+CD25+Foxp3+ Tregs in each group. Results are presented as mean±SEM of 18 mice from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
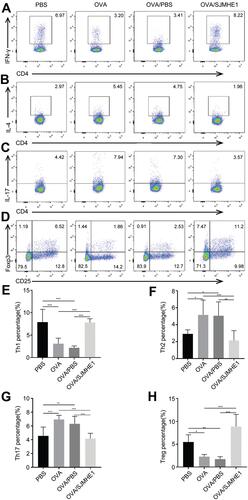
Figure 3 SJMHE1 treatment regulates the expression of transcription factor from lungs of allergic mice. On day 29, the mice were killed, and their lungs were tested for mRNA expression of T-bet (A), GATA3 (B), ROR-γt (C), and Foxp3 (D) by qRT-PCR. Results are presented as mean±SEM of 18 mice from three independent experiments performed in triplicate wells. *P < 0.05, ***P < 0.001.
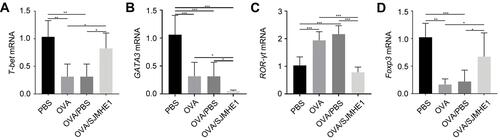
Figure 4 SJMHE1 treatment regulates the expression of cytokines from lungs of allergic mice. On day 29, the mice were killed, and their lungs were tested for mRNA expression of IL-4 (A), IL-5 (B), IL-13 (C), IFN-γ (D), IL-17 (E), IL-10 (F), TGF-β (G), and IL-35 (H) by qRT-PCR. Results are presented as mean±SEM of 18 mice from three independent experiments performed in triplicate wells. *P < 0.05, **P < 0.01, ***P < 0.001.
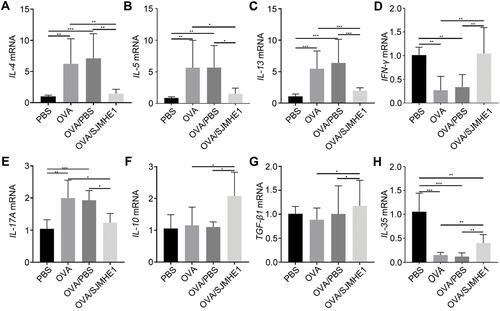
SJMHE1 Treatment Modulates miR-155 Expression, Which is Related to Th17/Treg Balance in Asthmatic Mice
miR-155 regulates the balance of Th1/Th2 and is a novel target for asthma,Citation26 while SJMHE1 effectively modulates Th cell distribution to suppress inflammatory response in asthmatic mice based on the above results. However, Th17/Treg imbalance is far beyond the Th1/Th2 imbalance and is involved in the development and severity of asthma.Citation33 Thus, we further investigated the miR-155 expression after SJMHE1 treatment and whether SJMHE1 regulates Th17/Treg balance through miR-155 in asthmatic mice. As expected, the level of miR-155 in the lungs of OVA or OVA/PBS-treated mice substantially increased compared with PBS-treated mice (). However, the expression of miR-155 was downregulated upon SJMHE1 treatment (). Moreover, miR-155 expression was positively correlated with Th17/Treg ratio in asthmatic mice (). Consistently, miR-155 expression was positively correlated with ROR-γt mRNA levels but negatively correlated with Foxp3 mRNA levels in asthmatic mice (). Therefore, SJMHE1 possibly regulates Th17/Treg cell balance by modulating miR-155 expression.
Figure 5 SJMHE1 treatment regulates the expression of miR-155 in lungs of allergic mice. On day 29, the mice were killed, and their lungs were tested for miR-155 expression by qRT-PCR. (A) miR-155 relative expression. Results are presented as mean±SEM of 12 mice from two independent experiments. (B) Correlation between miR-155 expression level and the ratio of Th17/Treg cells (n=6). (C) Correlation of miR-155 expression with that of ROR-γt and Foxp3 mRNA (D) in the lungs of OVA-treated mice (n=6). Spearman correlation analysis was used for the correlation calculation. ***P < 0.001.
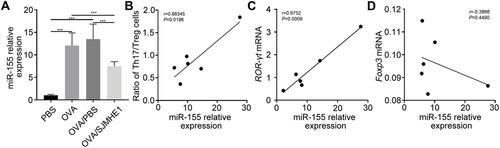
SJMHE1 Regulated STAT3 and STAT5 Activation in Lungs of Asthmatic Mice
STAT3 activates miR-155 in Th17 cells and promotes experimental autoimmune uveitis.Citation34 Furthermore, miR-155 is involved in the development and functional regulation of Treg in conventional T cells.Citation35 STAT3 and STAT5 are critical for mediating Th17 and Treg cell differentiation. IL-2 activates STAT5 to promote Treg cell differentiation, and IL-6 promotes Th17 differentiation via STAT3 activation.Citation36 SJMHE1 modulates the Th17 and Treg cell differentiation in asthmatic mice. Thus, we determined whether SJMHE1 regulates the expression of miR-155 and its effects on STAT3 and STAT5 activation in asthmatic mice. As expected, the expression of p-STAT3 in the lungs of OVA or OVA/PBS-treated mice was upregulated compared with PBS-treated mice, whereas p-STAT5 was downregulated (). However, SJMHE1 treatment enhanced the phosphorylation of STAT5 but suppressed p-STAT3 expression (). Moreover, the negative regulator SOCS1, which is miR-155 target, was downregulated in the lungs of OVA- or OVA/PBS-treated mice compared with PBS-treated mice (). However, SOCS1 expression was upregulated by SJMHE1 treatment (). Therefore, SJMHE1 may modulate Th17 and Treg differentiation by regulating STAT3 and STAT5 activation and inhibit the inflammatory response in asthmatic mice.
Figure 6 SJMHE1 treatment regulates the activation of STAT3 and STAT5 in the lungs of allergic mice. On day 29, the mice were killed, and their lungs were tested for p-STAT3 (Tyr705), p-STAT5 (Tyr694), and SOCS1 expression by Western blot analysis. Data are representative of two independent experiments.
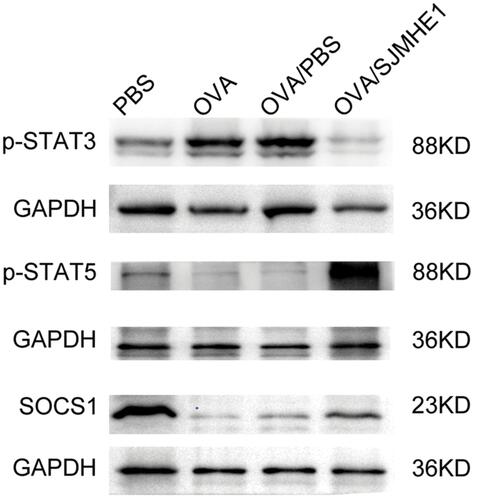
SJMHE1 Regulated Th17 and Treg Differentiation via miR-155 in vitro
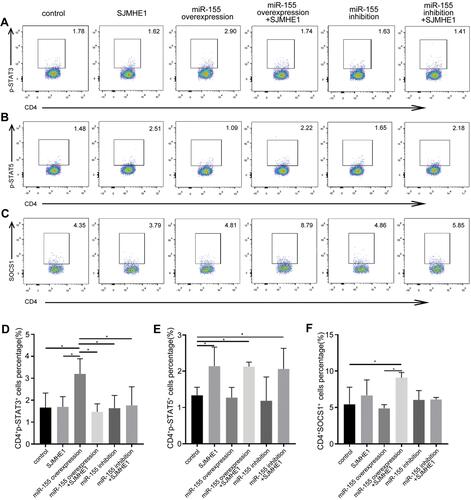
miR-155 could regulate Th17 and/or Treg response in various diseases,Citation29,Citation37 including asthma.Citation38 Thus, we further explored whether SJMHE1 could modulate the balance of Th17 and Treg of asthma via miR-155 in vitro. Mouse CD4+ T cells were transfected by a lentivirus containing over-expressed and knocked down miR-155, and then stimulated with MACSiBead particles in the presence or absence of SJMHE1. miR-155 expression significantly increased in CD4+ T cells transfected with lentivirus encoding miR-155 (overexpression), whereas no change was observed in cells transfected with miR-155 sponge (inhibition, Figure S1). As expected, the over-expression of miR-155 increased the proportion of Th17 cells in CD4+ T cells, but the proportion of Th17 cells decreased in the presence of SJMHE1 (). In comparison with the vector control, the proportion of Treg had no significant change in the over-expression or inhibition of miR-155 group. However, the addition of SJMHE1 increased the proportion of Treg regardless of the over-expression or inhibition of miR-155 (). In general, STAT3 and STAT5 signaling is critical for Th17 and Treg differentiation, respectively. Consistent with the findings on Th17 and Treg cells, the proportion of CD4+p-STAT3+ increased after transfecting lentivirus with over-expressed miR-155, but treatment with SJMHE1 reduced the expression of p-STAT3 in CD4+ T cells overexpressed miR-155, while that of p-STAT5 was elevated in CD4+ T cells after SJMHE1 treatment regardless over-expressed or inhibited miR-155 (). In comparison with vector control or over-expressed miR-155, SJMHE1 treatment increased the percentage of CD4+SOCS1+ cells (). Therefore, miR-155 promotes Th17 response, but SJMHE1 inhibits the miR-155-mediated Th17 cell response and promotes the induction of Treg bias in CD4+ T cells in vitro by regulating the activation of STAT3 and STAT5.
Figure 7 SJMHE1 regulates Th17 and Treg differentiation via miR-155 in vitro. CD4+ T cells were transfected with lentivirus containing over-expressed and knocked down miR-155, and then stimulated with MACSiBead particles in the presence or absence of 1 µg/mL SJMHE1 for 96 h. The cells were tested for Th17 and Treg subsets by flow cytometry. (A) CD4+IL-17+ Th17 cells and (B) CD4+CD25+Foxp3+ Tregs in each group are shown. Data are representative of three experiments. (C) Proportion of CD4+IL-17+ Th17 cells and (D) CD4+CD25+Foxp3+ Tregs in each group. Results are presented as mean±SEM of three experiments performed in triplicate. *P < 0.05, **P < 0.01.
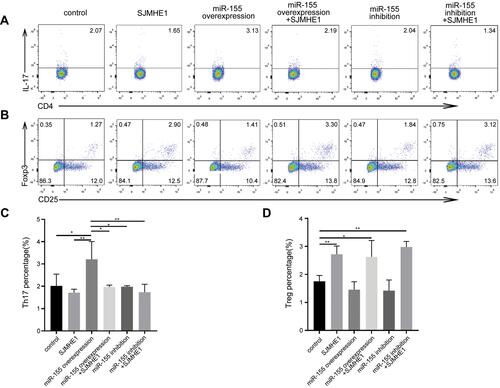
Figure 8 SJMHE1 regulates p-STAT3 and p-STAT5 expression via miR-155 in vitro. CD4+ T cells transfected with lentivirus containing over-expressed and knocked down miR-155, and then stimulated with MACSiBead particles in the presence or absence of 1 µg/mL SJMHE1 for 96 h. The cells were tested for p-STAT3, p-STAT5, and SOCS1 subsets by flow cytometry. (A) CD4+p-STAT3+ cells, (B) CD4+p-STAT5+ cells, and (C) CD4+SOCS1+ cells in each group. Data are representative of three experiments. (D) Percentage of CD4+p-STAT3+ cells, (E) CD4+p-STAT5+ Tregs, and (F) CD4+SOCS1+ cells in each group. Results are presented as mean±SEM of three experiments performed in triplicate. *P < 0.05.
Discussion
Asthma is an uncontrolled airway inflammatory response characterized by immune imbalance, especially when the imbalance of CD4+ T cells plays a critical role in the pathogenesis of asthma. Drug intervention can correct the imbalance of CD4+ T cell and improve respiratory symptoms in asthma.Citation39,Citation40 Interestingly, helminth parasite can balance between immune cell activation through co-evolving with the host, thus introducing a new pathway to control untoward immune responses, including asthma.Citation8 Our previous research found that the schistosoma peptide SJMHE1 can inhibit airway inflammation in asthmatic mice.Citation21 SJMHE1-treated macrophages and dendritic cells (DCs) show a tolerogenic phenotype and induce CD4+CD25+Foxp3+ Treg cell differentiation.Citation18 Although innate lymphoid cells, macrophages, eosinophils, mast cells, and T and B cells play an important role in the pathogenesis of asthma, the type 2 response is usually the hallmark of asthma.Citation41 The Th17 cell response has been related to the severity of asthma,Citation42 and high levels of IL-17 indicate resistance to corticosteroid treatment in asthmatic mice and patients.Citation43,Citation44 Thus, in the current study, we evaluated the effect of SJMHE1 on Th17 and Treg cells in asthmatic mice and demonstrated that SJMHE1 inhibits the development of asthma by regulating the activation of STAT3 and STAT5 via miR-155, thereby modulating the balance of Th17/Treg.
Similar to the intervention with SJMHE1 at the beginning of OVA-induced asthma,Citation21 SJMHE1 still relieved the development of asthma, reduced the infiltration of inflammatory cells in the lungs, and decreased the total cell, eosinophilia, and neutrophil number in the BALF of mice, which were treated by SJMHE1 from the third OVA sensitization. SJMHE1 treatment upregulated the proportion of Th1 and Treg cells and downregulated the proportion of Th2 and Th17 cells in the lungs of asthmatic mice. Consistent with the regulation of Th cells by SJMHE1, SJMHE1 treatment upregulated the expression of T-bet and Foxp3 mRNA and downregulated the expression of GATA3 and ROR-γt mRNA of lungs in asthmatic mice. Consistent with our previous study,Citation21 SJMHE1 treatment reduced the expression of IL-4, IL-5, and IL-13 mRNA and increased the expression of IFN-γ, IL-10, TGF-β, and IL-35 mRNA in the lungs of asthmatic mice. Treg cells usually suppress inflammatory responses via the production of IL-10, IL-35, and TGF-β.Citation13 The increase in IFN-γ, IL-10, TGF-β, and IL-35 mRNA expression was consistent with the increase in Th1 and Treg cell proportions after SJMHE1 treatment in asthmatic mice. SJMHE1 regulated the balance of Th cells and cytokines in the lungs of asthmatic mice and may offer protection against the development of asthma. However, how SJMHE1 regulates the balance of Th cells remains unknown.
miRNA-mediated controlled gene expression can fine-tune the appropriate immune response, which is essential in helminth infection and immunity.Citation45 The excretory/secretory (ES) antigens of Taenia crassiceps can inhibit LPS-induced Let-7i in human dendritic cells (DCs) related to the decrease in inflammatory response.Citation46 Furthermore, miRNAs participate in the regulation of T cell polarization and cytokine production. miR-155 was initial thought as a “Th2 enriched” miRNA, which was essential in mediating allergy and antihelminth immunity.Citation47 Recent studies show that miR-155 is involved in controlling Treg function and Th17-mediated inflammation.Citation35,Citation48 Furthermore, miR-155 is a critical target in asthma, and the upregulation of miR-155 participates in the development of asthma.Citation26,Citation30 Whether miR-155 is involved in the SJMHE1 regulating Th cell balance in asthmatic mice has not been determined. Consistent with the upregulation of miR-155 expression in asthmatic mice,Citation30 this study also demonstrated higher miR-155 expression in OVA and OVA/PBS groups than PBS control. However, SJMHE1 treatment significantly inhibited the expression of miR-155 in lungs of asthmatic mice (). These results are consistent with a previous publication in which miR-155 deficiency reduced the Th2 cell and Th2 cytokine levels and contributed to the alleviation of airway inflammation in asthmatic mice.Citation30 Th17/Treg imbalance is far beyond Th1/Th2 imbalance,Citation33 and whether miR-155 regulates Th17/Treg balance in asthma remains unclear. The present study focused on SJMHE1 regulating Th17/Treg balance in asthmatic mice through miR-155. Indeed, miR-155 expression was positively correlated with Th17/Treg ratio and RORγt mRNA levels but negatively correlated with Foxp3 mRNA levels in asthmatic mice (). SJMHE1 might regulate Th17/Treg balance through miR-155 in asthmatic mice.
In general, STAT signal is critical for Th17/Treg cell differentiation. IL-6/STAT3 is essential for Th17 cell differentiation, while IL-2/STAT5 is indispensable for Treg cells.Citation49,Citation50 To explore the regulation of Th17/Treg cells by SJMHE1, we tested the expression of p-STAT3 and p-STAT5 in lungs of mice. Consistent with the proportion of Th17 and Treg cells in asthmatic mice, OVA and OVA/PBS groups showed the upregulation of p-STAT3 and downregulation of p-STAT5. However, OVA/SJMHE1 group showed the upregulation of p-STAT5 and downregulation of p-STAT3. Moreover, consistent with miR-155 expression, as a target of miR-155 and an important negative regulator of JAK/STAT signal, SOCS1 expression was downregulated in OVA and OVA/PBS groups but upregulated in OVA/SJMHE1 (). SOCS1 negatively regulated the Th17 cell differentiation by blocking the IL-6-induced activation of STAT3.Citation51,Citation52 This finding supports that miR-155 regulates Th17 and Treg cell differentiation by targeting SOCS1.Citation29,Citation48 SJMHE1 might regulate Th17/Treg balance by regulating the activation of STAT3 and STAT5, thereby ameliorating the airway inflammation of asthmatic mice.
To confirm the regulation of Th17/Treg cells by SJMHE1 via miR-155, we stimulated mouse CD4+ T cells after forcing the expression or inhibition of miR-155 by lentivirus. Indeed, the over-expression of miR-155 increased the frequency of Th17 cells. However, in the presence of SJMHE1, the proportion of Th17 cells decreased (). Although the over-expression or inhibition of miR-155 did not increase the proportion of Treg cells, with the addition of SJMHE1, the frequency of Treg cells increased (). Consistent with the proportion of Th17 and Treg cells, the over-expression of miR-155 increased the frequency of CD4+p-STAT3+ cells. The addition of SJMHE1 reduced the proportion of CD4+p-STAT3+ cells increased the frequency of CD4+p-STAT5+ cells regardless of the over-expression or inhibition of miR-155 (). Consistent with the increase in SOCS1 expression in OVA/SJMHE1-treated mice, SJMHE1 treatment increased the proportion of CD4+SOCS1+ cells in vitro (). The increase of SOCS1+ cells is consistent with the decrease of p-STAT3+ cells in CD4+ T cells after SJMHE1 treatment. Nevertheless, another publication showed that the over-expression of miR-155 by pre-miR-155 increased the frequencies of Th17 and Treg cells, and the expression of p-STAT3 and p-STAT5, but the inhibition of miR-155 by anti-miR-155 reduced the proportion of Th17 and Treg cells and the expression of p-STAT3 and p-STAT5 in CD4+ T cells.Citation48 This discrepancy is associated with the difference in miR-155 expression system or CD4+ T cell treatment method, thus requiring further investigation. Our results are consistent with another publication in which JAK2 inhibitor AG490 regulates the balance of Th17 and Treg cells by modulating the activation of STAT3 and STAT5 in autoimmune arthritis mice. A490 treatment reduced the proportion of Th17 cells and the expression of p-STAT3 but increased the proportions of Treg cells and the expression of p-STAT5.Citation53 A reciprocal regulation of STAT3 and STAT5 was observed under an inflammatory condition. Transfection with STAT3 siRNA in CD4+ T cells inhibited Th17 cell differentiation but increased the frequency of Treg cells from rheumatoid arthritis (RA) peripheral blood and synovial fluid. Similarly, the suppression of STAT5 decreased Treg cells and increased Th17 cells.Citation54 Furthermore, the results of our previous and the present study support that reciprocal activation was observed between STAT3 and STAT5.Citation55–Citation57 However, the mechanism of reciprocal regulation of STAT3 and STAT5 needs to be explored. Therefore, SJMHE1 regulates Th17 and Treg balance by regulating the activation of STAT3 and STAT5 via miR-155 in vitro.
Conclusion
In conclusion, we confirmed that SJMHE1 ameliorated ongoing asthma by regulating the balance of Th cells. SJMHE1 suppressed the activation of STAT3 and upregulated the activation of STAT5 via miR-155, thereby regulating the balance of Th17 and Treg cells in asthma. The schematic representation of SJMHE1 regulation of Th17/Treg balance in asthma is shown in . Our results further confirmed that S. japonicum peptide SJMHE1 might be a promising treatment for asthma.
Ethics Approval
Mouse handling and experimentation were conducted in strict compliance with the Regulations for Administration of Affairs Concerning Experimental Animals of Jiangsu University. All protocols were approved by the Jiangsu University Institutional Animal Care and Use Committee (Permit Number: JSU 18–012).
Disclosure
The authors declare that they have no competing interests.
Additional information
Funding
References
- Xue F, Yu M, Li L, et al. Elevated granulocytic myeloid-derived suppressor cells are closely related with elevation of Th17 cells in mice with experimental asthma. Int J Biol Sci. 2020;16(12):2072–2083. doi:10.7150/ijbs.43596
- Muehling LM, Lawrence MG, Woodfolk JA. Pathogenic CD4(+) T cells in patients with asthma. J Allergy Clin Immunol. 2017;140(6):1523–1540. doi:10.1016/j.jaci.2017.02.025
- Shi YH, Shi GC, Wan HY, et al. Coexistence of Th1/Th2 and Th17/Treg imbalances in patients with allergic asthma. Chin Med J (Engl). 2011;124(13):1951–1956.
- Corren J, Lemanske RF, Hanania NA, et al. Lebrikizumab treatment in adults with asthma. N Engl J Med. 2011;365(12):1088–1098. doi:10.1056/NEJMoa1106469
- Just J, Deschildre A, Lejeune S, Amat F. New perspectives of childhood asthma treatment with biologics. Pediatr Allergy Immunol. 2019;30(2):159–171.
- Maizels RM, McSorley HJ. Regulation of the host immune system by helminth parasites. J Allergy Clin Immunol. 2016;138(3):666–675. doi:10.1016/j.jaci.2016.07.007
- Gazzinelli-Guimaraes PH, Nutman TB. Helminth parasites and immune regulation. F1000Res. 2018;7:F1000 Faculty Rev–1685. doi:10.12688/f1000research.15596.1
- Maizels RM. Regulation of immunity and allergy by helminth parasites. Allergy. 2020;75(3):524–534. doi:10.1111/all.13944
- Fernandes JS, Cardoso LS, Pitrez PM, Cruz AA. Helminths and asthma: risk and protection. Immunol Allergy Clin North Am. 2019;39(3):417–427. doi:10.1016/j.iac.2019.03.009
- Bohnacker S, Troisi F, de Los Reyes Jimenez M, Esser-von Bieren J. What can parasites tell us about the pathogenesis and treatment of asthma and allergic diseases. Front Immunol. 2020;11:2106. doi:10.3389/fimmu.2020.02106
- Pacifico LG, Marinho FA, Fonseca CT, et al. Schistosoma mansoni antigens modulate experimental allergic asthma in a murine model: a major role for CD4+ CD25+ Foxp3+ T cells independent of interleukin-10. Infect Immun. 2009;77(1):98–107. doi:10.1128/IAI.00783-07
- Navarro S, Pickering DA, Ferreira IB, et al. Hookworm recombinant protein promotes regulatory T cell responses that suppress experimental asthma. Sci Transl Med. 2016;8(362):362ra143. doi:10.1126/scitranslmed.aaf8807
- Logan J, Navarro S, Loukas A, Giacomin P. Helminth-induced regulatory T cells and suppression of allergic responses. Curr Opin Immunol. 2018;54:1–6. doi:10.1016/j.coi.2018.05.007
- Amu S, Saunders SP, Kronenberg M, Mangan NE, Atzberger A, Fallon PG. Regulatory B cells prevent and reverse allergic airway inflammation via FoxP3-positive T regulatory cells in a murine model. J Allergy Clin Immunol. 2010;125(5):1114–1124e1118. doi:10.1016/j.jaci.2010.01.018
- Coltherd JC, Rodgers DT, Lawrie RE, et al. The parasitic worm-derived immunomodulator, ES-62 and its drug-like small molecule analogues exhibit therapeutic potential in a model of chronic asthma. Sci Rep. 2016;6(1):19224. doi:10.1038/srep19224
- Obieglo K, Schuijs MJ, Ozir-Fazalalikhan A, et al. Isolated Schistosoma mansoni eggs prevent allergic airway inflammation. Parasite Immunol. 2018;40(10):e12579. doi:10.1111/pim.12579
- Reece JJ, Siracusa MC, Southard TL, Brayton CF, Urban JF Jr., Scott AL. Hookworm-induced persistent changes to the immunological environment of the lung. Infect Immun. 2008;76(8):3511–3524. doi:10.1128/IAI.00192-08
- Wang X, Zhou S, Chi Y, et al. CD4+CD25+ Treg induction by an HSP60-derived peptide SJMHE1 from Schistosoma japonicum is TLR2 dependent. Eur J Immunol. 2009;39(11):3052–3065. doi:10.1002/eji.200939335
- Wang X, Wang J, Liang Y, et al. Schistosoma japonicum HSP60-derived peptide SJMHE1 suppresses delayed-type hypersensitivity in a murine model. Parasit Vectors. 2016;9(1):147. doi:10.1186/s13071-016-1434-4
- Wang X, Li L, Wang J, et al. Inhibition of cytokine response to TLR stimulation and alleviation of collagen-induced arthritis in mice by Schistosoma japonicum peptide SJMHE1. J Cell Mol Med. 2017;21(3):475–486. doi:10.1111/jcmm.12991
- Zhang W, Li L, Zheng Y, et al. Schistosoma japonicum peptide SJMHE1 suppresses airway inflammation of allergic asthma in mice. J Cell Mol Med. 2019;23(11):7819–7829. doi:10.1111/jcmm.14661
- Chen Y, Mao ZD, Shi YJ, et al. Comprehensive analysis of miRNA-mRNA-lncRNA networks in severe asthma. Epigenomics. 2019;11(2):115–131. doi:10.2217/epi-2018-0132
- Specjalski K, Jassem E. MicroRNAs: potential biomarkers and targets of therapy in allergic diseases? Arch Immunol Ther Exp (Warsz). 2019;67(4):213–223. doi:10.1007/s00005-019-00547-4
- Suojalehto H, Lindstrom I, Majuri ML, et al. Altered microRNA expression of nasal mucosa in long-term asthma and allergic rhinitis. Int Arch Allergy Immunol. 2014;163(3):168–178. doi:10.1159/000358486
- Xiao R, Noel A, Perveen Z, Penn AL. In utero exposure to second-hand smoke activates pro-asthmatic and oncogenic miRNAs in adult asthmatic mice. Environ Mol Mutagen. 2016;57(3):190–199. doi:10.1002/em.21998
- Zhou H, Li J, Gao P, Wang Q, Zhang J. miR-155: a novel target in allergic asthma. Int J Mol Sci. 2016;17(10):10. doi:10.3390/ijms17101773
- Banerjee A, Schambach F, DeJong CS, Hammond SM, Reiner SL. Micro-RNA-155 inhibits IFN-gamma signaling in CD4+ T cells. Eur J Immunol. 2010;40(1):225–231. doi:10.1002/eji.200939381
- Kohlhaas S, Garden OA, Scudamore C, Turner M, Okkenhaug K, Vigorito E. Cutting edge: the Foxp3 target miR-155 contributes to the development of regulatory T cells. J Immunol. 2009;182(5):2578–2582. doi:10.4049/jimmunol.0803162
- Wang D, Tang M, Zong P, et al. MiRNA-155 regulates the Th17/Treg ratio by targeting SOCS1 in severe acute pancreatitis. Front Physiol. 2018;9:686. doi:10.3389/fphys.2018.00686
- Malmhall C, Alawieh S, Lu Y, et al. MicroRNA-155 is essential for T(H)2-mediated allergen-induced eosinophilic inflammation in the lung. J Allergy Clin Immunol. 2014;133(5):1429–1438. doi:10.1016/j.jaci.2013.11.008
- Bergallo M, Accorinti M, Galliano I, et al. Expression of miRNA 155, FOXP3 and ROR gamma, in children with moderate and severe atopic dermatitis. G Ital Dermatol Venereol. 2020;155(2):168–172. doi:10.23736/S0392-0488.17.05707-8
- Xia G, Wu S, Wang X, Fu M. Inhibition of microRNA-155 attenuates concanavalin-A-induced autoimmune hepatitis by regulating Treg/Th17 cell differentiation. Can J Physiol Pharmacol. 2018;96(12):1293–1300. doi:10.1139/cjpp-2018-0467
- Wang L, Wan H, Tang W, et al. Critical roles of adenosine A2A receptor in regulating the balance of Treg/Th17 cells in allergic asthma. Clin Respir J. 2018;12(1):149–157. doi:10.1111/crj.12503
- Escobar T, Yu CR, Muljo SA, Egwuagu CE. STAT3 activates miR-155 in Th17 cells and acts in concert to promote experimental autoimmune uveitis. Invest Ophthalmol Vis Sci. 2013;54(6):4017–4025. doi:10.1167/iovs.13-11937
- Lu LF, Thai TH, Calado DP, et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30(1):80–91. doi:10.1016/j.immuni.2008.11.010
- Wei L, Laurence A, O’Shea JJ. New insights into the roles of Stat5a/b and Stat3 in T cell development and differentiation. Semin Cell Dev Biol. 2008;19(4):394–400. doi:10.1016/j.semcdb.2008.07.011
- Escobar TM, Kanellopoulou C, Kugler DG, et al. miR-155 activates cytokine gene expression in Th17 cells by regulating the DNA-binding protein Jarid2 to relieve polycomb-mediated repression. Immunity. 2014;40(6):865–879. doi:10.1016/j.immuni.2014.03.014
- Qiu L, Zhang Y, Do DC, et al. miR-155 modulates cockroach allergen- and oxidative stress-induced cyclooxygenase-2 in asthma. J Immunol. 2018;201(3):916–929. doi:10.4049/jimmunol.1701167
- Ding F, Fu Z, Liu B. Lipopolysaccharide exposure alleviates asthma in mice by regulating Th1/Th2 and Treg/Th17 balance. Med Sci Monit. 2018;24:3220–3229. doi:10.12659/MSM.905202
- Liang P, Peng S, Zhang M, Ma Y, Zhen X, Huai LH. Qi Huang corrects the balance of Th1/Th2 and Treg/Th17 in an ovalbumin-induced asthma mouse model. Biosci Rep. 2017;37(6):BSR20171071. doi:10.1042/BSR20171071
- Lambrecht BN, Hammad H. The immunology of asthma. Nat Immunol. 2015;16(1):45–56. doi:10.1038/ni.3049
- Allen JE, Sutherland TE, Ruckerl D. IL-17 and neutrophils: unexpected players in the type 2 immune response. Curr Opin Immunol. 2015;34:99–106. doi:10.1016/j.coi.2015.03.001
- Nanzer AM, Chambers ES, Ryanna K, et al. Enhanced production of IL-17A in patients with severe asthma is inhibited by 1alpha,25-dihydroxyvitamin D3 in a glucocorticoid-independent fashion. J Allergy Clin Immunol. 2013;132(2):297–304. doi:10.1016/j.jaci.2013.03.037
- McKinley L, Alcorn JF, Peterson A, et al. TH17 cells mediate steroid-resistant airway inflammation and airway hyperresponsiveness in mice. J Immunol. 2008;181(6):4089–4097. doi:10.4049/jimmunol.181.6.4089
- Entwistle LJ, Wilson MS. MicroRNA-mediated regulation of immune responses to intestinal helminth infections. Parasite Immunol. 2017;39(2):e12406. doi:10.1111/pim.12406
- Terrazas LI, Sanchez-Munoz F, Perez-Miranda M, et al. Helminth excreted/secreted antigens repress expression of LPS-induced Let-7i but not miR-146a and miR-155 in human dendritic cells. Biomed Res Int. 2013;2013:972506. doi:10.1155/2013/972506
- Okoye IS, Czieso S, Ktistaki E, et al. Transcriptomics identified a critical role for Th2 cell-intrinsic miR-155 in mediating allergy and antihelminth immunity. Proc Natl Acad Sci U S A. 2014;111(30):E3081–3090. doi:10.1073/pnas.1406322111
- Yao R, Ma YL, Liang W, et al. MicroRNA-155 modulates Treg and Th17 cells differentiation and Th17 cell function by targeting SOCS1. PLoS One. 2012;7(10):e46082. doi:10.1371/journal.pone.0046082
- Wan CK, Andraski AB, Spolski R, et al. Opposing roles of STAT1 and STAT3 in IL-21 function in CD4+ T cells. Proc Natl Acad Sci U S A. 2015;112(30):9394–9399. doi:10.1073/pnas.1511711112
- Zorn E, Nelson EA, Mohseni M, et al. IL-2 regulates FOXP3 expression in human CD4+CD25+ regulatory T cells through a STAT-dependent mechanism and induces the expansion of these cells in vivo. Blood. 2006;108(5):1571–1579. doi:10.1182/blood-2006-02-004747
- Yu CR, Mahdi RR, Oh HM, et al. Suppressor of cytokine signaling-1 (SOCS1) inhibits lymphocyte recruitment into the retina and protects SOCS1 transgenic rats and mice from ocular inflammation. Invest Ophthalmol Vis Sci. 2011;52(9):6978–6986.
- Jager LD, Dabelic R, Waiboci LW, et al. The kinase inhibitory region of SOCS-1 is sufficient to inhibit T-helper 17 and other immune functions in experimental allergic encephalomyelitis. J Neuroimmunol. 2011;232(1–2):108–118. doi:10.1016/j.jneuroim.2010.10.018
- Park JS, Lee J, Lim MA, et al. JAK2-STAT3 blockade by AG490 suppresses autoimmune arthritis in mice via reciprocal regulation of regulatory T cells and Th17 cells. J Immunol. 2014;192(9):4417–4424. doi:10.4049/jimmunol.1300514
- Ju JH, Heo YJ, Cho ML, et al. Modulation of STAT-3 in rheumatoid synovial T cells suppresses Th17 differentiation and increases the proportion of Treg cells. Arthritis Rheum. 2012;64(11):3543–3552. doi:10.1002/art.34601
- Dong L, Wang X, Tan J, et al. Decreased expression of microRNA-21 correlates with the imbalance of Th17 and Treg cells in patients with rheumatoid arthritis. J Cell Mol Med. 2014;18(11):2213–2224. doi:10.1111/jcmm.12353
- Park JS, Kwok SK, Lim MA, et al. STA-21, a promising STAT-3 inhibitor that reciprocally regulates Th17 and Treg cells, inhibits osteoclastogenesis in mice and humans and alleviates autoimmune inflammation in an experimental model of rheumatoid arthritis. Arthritis Rheumatol. 2014;66(4):918–929. doi:10.1002/art.38305
- Jhun JY, Moon SJ, Yoon BY, et al. Grape seed proanthocyanidin extract-mediated regulation of STAT3 proteins contributes to Treg differentiation and attenuates inflammation in a murine model of obesity-associated arthritis. PLoS One. 2013;8(11):e78843. doi:10.1371/journal.pone.0078843