Abstract
Background
The Wnt pathway plays an important role in Hepatocarcinogenesis. We analyzed the association of the Wnt pathway with the proliferation of hepatoma cells using Wnt3a and niclosamide, a drug used to treat tapeworm infection.
Methods
We performed an MTS assay to determine whether Wnt3a stimulated proliferation of Huh-6 and Hep3B human hepatoma cell lines after 72 hours of incubation with Wnt3a in serum-free medium. The cells were subjected to hematoxylin and eosin staining and terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) after 48 hours of incubation. RNA was isolated 48 hours after addition of Wnt3a or niclosamide, and cyclin D1 expression levels were analyzed by real-time quantitative polymerase chain reaction. The promoter activity of T-cell factor was analyzed by luciferase assay 48 hours after transfection of TOPflash. Western blot analysis was performed with antibodies against β-catenin, dishevelled 2, and cyclin D1.
Results
Cell proliferation increased with Wnt3a. Niclosamide suppressed proliferation with or without Wnt3a. Hematoxylin and eosin and TUNEL staining suggested that apoptosis occurred in cells with niclosamide. Cyclin D1 was upregulated in the presence of Wnt3a and downregulated with addition of niclosamide. The promoter activity of T-cell factor increased with Wnt3a, whereas T-cell factor promoter activity decreased with niclosamide. Western blot analysis showed that Wnt3a upregulated β-catenin, dishevelled 2, and cyclin D1, while niclosamide downregulated them.
Conclusion
Niclosamide is a potential candidate for the treatment of hepatoma.
Introduction
Molecular therapy is a promising treatment for hepatoma. Currently, molecular therapy targets signaling pathways that are aberrantly activated specifically in cancer cells. The Wnt signaling pathway has been found to be dysregulated in hepatoma, as revealed by comparison of expression levels of Frizzled (Fz), the receptor for Wnt protein, between cancerous and noncancerous tissues.Citation1 Based on these results, the Wnt signaling pathway is expected to be a target of molecular therapy for hepatoma.
The Wnt signaling pathway plays a crucial role in regulating cell growth and differentiation. Cytoplasmic β-catenin is maintained at low levels in the absence of Wnt signals through destruction by the glycogen synthase kinase-3 β complex.Citation2 Wnt proteins bind to their receptor, Fz, and coreceptor, low-density lipoprotein receptor-related protein 5 and 6, to form a complex.Citation3 The complex phosphorylates low-density lipoprotein receptor-related protein 5 and 6 and sequesters the axin-adenomatous polyposis coli-dishevelled (DVL) destruction complex to the receptor domain, inhibiting glycogen synthase kinase-3β. β-catenin then accumulates in the cytosol and translocates to the nucleus. In the nucleus, β-catenin acts as a cofactor of T-cell factor (TCF)/lymphoid enhancer factor and activates target genes. Constitutive activation of the Wnt pathway leads to abnormal cell growth and development of cancer.Citation4 The Fz family of genes, which consists of 10 members, function as cell surface receptors.Citation5 Fz9 is widely expressed in hepatoma cell lines, and its inhibition suppresses hepatoma cell proliferation.Citation6
Niclosamide was originally developed for the treatment of tapeworm infections, and is still being used for this purpose worldwide;Citation7,Citation8 however, its pharmacologic mechanism is not well understood. Among the number of chemicals screened, niclosamide was found to cause autophagy and reversibly inhibit the mammalian target of rapamycin complex 1.Citation9,Citation10 Sack et al showed that niclosamide exerts a suppressive effect on the metastasis-promoting activity of S100-A4, a calcium-binding protein that promotes colon cancer metastasis.Citation11 In addition, they demonstrated that niclosamide suppresses cell proliferation and metastasis, and induces apoptosis of colorectal cancer cells.Citation11 Chen et al proposed that niclosamide induces internalization of Fz1.Citation12 Using an established cell line, Fzd1GFP-U2OS, which stably expresses Fz1 labeled with green fluorescent protein, they found the green fluorescent protein signal to be internalized in cells incubated with niclosamide. Downregulation of DVL2 is another mechanism by which niclosamide inhibits the Wnt pathway.Citation13 Further, Yo et al screened 1,200 clinically approved drugs for drug-resistant ovarian cancer cells to identify niclosamide-disrupting metabolic pathways.Citation14 These reports suggest that niclosamide is a potential anticancer agent.
The Wnt pathway is activated in human hepatoma. Wnt3a, Wnt4a, and Wnt5a, as well as Fz3, Fz6, and Fz7, are upregulated in human hepatoma tissuesCitation1 when compared with surrounding normal tissues. Fz1 is also upregulated, although not as frequently as Fz3, Fz6, and Fz7. Fujimoto et al analyzed the expression of Fz in the human fetal and adult liver and in seven hepatoma cell lines with reverse transcriptase polymerase chain reaction (PCR).Citation6 They found that Fz1 is expressed in three cell lines (HLF, PLC/PRL/5, and Huh-6) and in the fetal liver, but not in the adult liver. These reports indicate that Wnt ligands act as growth factors and are inhibited by niclosamide in hepatoma.
Materials and methods
Cell culture
In this study, Huh-6 cells and Hep3B cells were purchased from RIKEN Cell Bank (Tsukuba, Japan) because the former expresses Fz1 and the latter does not.Citation6 Both cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (Sigma-Aldrich, St Louis, MO, USA) supplemented with 10% fetal bovine serum (Life Technologies, Grand Island, NY, USA). The cells were cultured with 5% carbon dioxide at 37°C in a humidified chamber. Cells were spread onto four-well chamber slides (Becton Dickinson, Franklin Lakes, NJ, USA) and subjected to hematoxylin and eosin staining and terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL). The stained slides were observed under an AX80 microscope (Olympus, Tokyo, Japan).
Cell proliferation analysis
Huh-6 cells were trypsinized, harvested, and spread onto 96-well flat-bottom plates (Asahi Techno Glass, Tokyo, Japan) at a density of 1,000 cells per well, and then incubated for 24 hours in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum. After culture, the cells were treated with Wnt3a (R&D Systems, Minneapolis, MN, USA) or niclosamide (Sigma-Aldrich) at the indicated concentrations for 72 hours. The cell cultures were then examined by 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium inner salt (MTS) according to the manufacturer’s instructions (Promega Corporation, Tokyo, Japan). MTS is bioreduced by cells into a colored formazan product that reduces absorbance at 490 nm. Absorbance was analyzed at a wavelength of 490 nm with an iMark Microplate Absorbance Reader (Bio-Rad, Hercules, CA, USA).
TUNEL staining
Apoptotic cells were detected using an apoptosis in situ detection kit (Wako Pure Chemicals, Osaka, Japan). Analysis of apoptotic cells was based on the TUNEL procedure, which consists of the addition of apoptotically fragmented DNA to the 3′ termini by terminal deoxynucleotidyl transferase followed by immunochemical detection using an anti-fluorescein antibody conjugated with horseradish peroxidase and diaminobenzidine as a substrate.
Real-time quantitative PCR
Cells were spread onto 6-well plates (Asahi Techno Glass) and cultured. After 48 hours of treatment with Wnt3a and niclosamide, total RNA (5 μg) was isolated with Isogen (Nippon Gene, Tokyo, Japan) and used to generate cDNA with SuperScript III and oligo(dT) primers according to the manufacturer’s instructions (Life Technologies). The PCR primers and product sizes were as follows: cyclin D1 (NM_053056, 5′-AGAGGCGGAGGAGAACAAACAG, 5′-AGGCGGTAGTAGGACAGGAAGTTG; 180 bp) and RPL19 (BC095445, 5′-CGAATGCCAGAGAAGGTCAC, 5′-CCATGAGAATCCGCTTGTTT; 157 bp). Real-time quantitative PCR was performed with 40 cycles of 5 seconds of denaturation and 5 seconds of annealing extension. RPL19 was used as an internal control.
Luciferase assay
Huh-6 cells were spread onto 24-well plates (Asahi Techno Glass) and cultured for 24 hours. When cells reached 70% confluence, they were transfected with Lipofectamine LTX (Life Technologies), 0.5 μg of TOPflash reporter plasmid (Millipore, Temecula, CA, USA), and 0.05 μg of pRL-TK (Promega, Madison, WI, USA) in the medium. Transcriptional activity was measured with a dual luciferase reporter assay system (Promega) using Gene Light (GL-200A) (Microtech Co Ltd, Funabashi, Japan). Plasmid in medium without transfection was used as a negative control.
Western blot analysis
Protein was isolated from cells after 48 hours of culture. A 10 μg sample of protein was subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis, and transferred to a nylon filter. After 30 minutes of incubation with 5% skim milk, the filters were incubated with primary antibodies for one hour at room temperature (1:2,500). After one hour of incubation with secondary antibodies at room temperature (1:2,500), the specific antigen-antibody complexes were visualized by enhanced chemiluminescence (GE Healthcare, Pittsburg, PA, USA). Rabbit monoclonal anti-β-catenin antibody and anti-DVL2 antibody were purchased from Cell Signaling Technologies (Danvers, MA, USA). Rabbit polyclonal cyclin D1 antibody and mouse monoclonal anti-tubulin-α antibody were purchased from Epitomics Inc (Burlingame, CA, USA) and Lab Vision (Fremont, CA, USA), respectively. Horseradish peroxidase-linked anti-rabbit antibody and horseradish peroxidase-linked anti-mouse antibody were purchased from GE Healthcare. The filter was reprobed with anti-tubulin-α antibody. Expression levels of β-catenin, DVL2, and cyclin D1 were normalized with tubulin-α and analyzed using ImageJ64 imaging software (National Institutes of Health, Bethesda, MD, USA).
Statistical analysis
Cell proliferation and real-time quantitative PCR data were analyzed by one-factor analysis of variance. Statistical analysis was performed using JMP5.0 J software (SAS Institute Japan, Tokyo, Japan). P<0.05 was accepted as being statistically significant.
Results
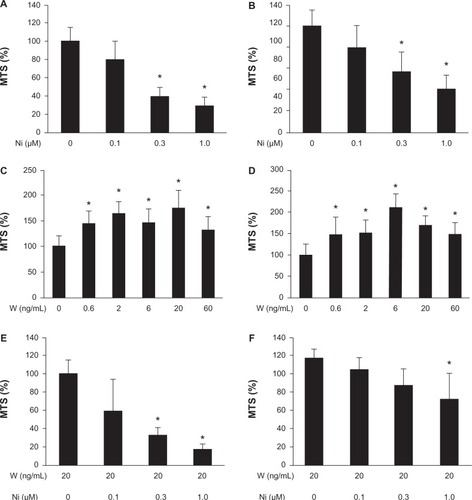
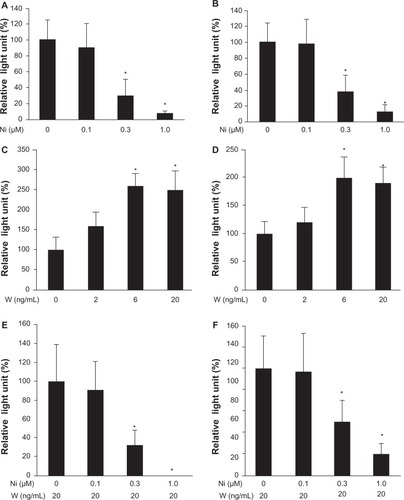
We performed the MTS assay to determine whether niclosamide suppressed proliferation of Huh-6 cells and Hep3B cells. Both cell lines were incubated with niclosamide for 72 hours. Serum was not added to the medium to quench its effects and focus on the effects of Wnt3a or niclosamide.Citation15 Cell proliferation was suppressed to 29%±9.0% in Huh-6 cells (P<0.05, ) and 41%±11% in Hep3B cells (P<0.05, ) at 1 μM of niclosamide. We performed the MTS assay to analyze whether Wnt3a stimulated proliferation of Huh-6 cells and Hep3B cells. Cell proliferation increased to 175%±33% in Huh-6 cells (P<0.05, ) and 170%±20% in Hep3B cells (P<0.05, ) with 20 ng/mL Wnt3a, respectively. Niclosamide was added to medium containing 20 ng/mL Wnt3a to determine if this suppressed the proliferative effect of Wnt3a. The MTS assay was performed 72 hours after addition of niclosamide at the indicated concentration. Niclosamide at 1.0 μM suppressed proliferation of Huh-6 cells to 17.0%±6% (P<0.05, ) and Hep3B cells to 62%±23% (P<0.05, ). These data indicate that niclosamide inhibits the Wnt3a-mediated proliferative effect.
Figure 1 Cell proliferation assay. A cell proliferation assay was performed after addition of Wnt3a or niclosamide to huh-6 cells (A, C, and E) and Hep3B cells (B, D, and F). Niclosamide suppressed proliferation without Wnt3a (A and B) or with Wnt3a (E and F). Wnt 3a increased proliferation (C and D).
Abbreviations: W, Wnt3a; Ni, niclosamide; MTS, 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium inner salt.
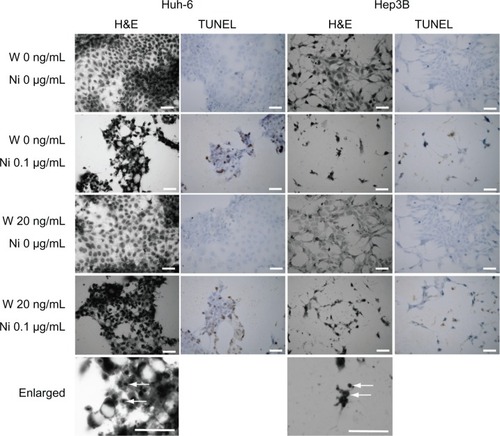
The cells were stained with hematoxylin and eosin for morphologic analysis (). We observed no changes in cells cultured in serum-free medium containing 20 ng/mL Wnt3a. On the other hand, Huh-6 cells and Hep3B cells showed morphologic changes after addition of 0.1 μM niclosamide, some with pyknotic nuclei suggesting apoptotic cell death (arrows). TUNEL staining was performed to confirm apoptosis. Positive results were observed in Huh-6 cells and Hep3B cells treated with niclosamide alone or with Wnt3a and niclosamide.
Figure 2 H&E staining and TUNEL staining. Huh-6 and Hep3B cells were subjected to H&E and TUNEL staining. While no change was seen in cells cultured without niclosamide cells, those treated with 0.1 μM niclosamide were damaged and had pyknotic nuclei (enlarged, white arrows).
Abbreviations: W, Wnt3a; Ni, niclosamide; TUNEL, terminal deoxynucleotidyl transferase (TdT)-mediated dUTP nick end-labeling; H&E, hematoxylin and eosin.
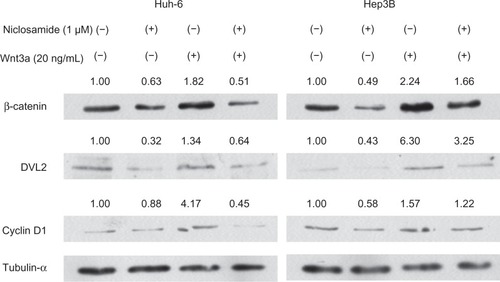
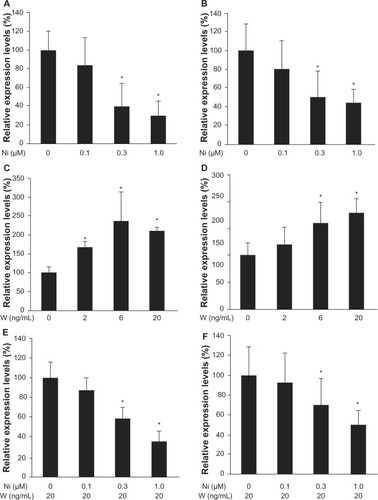
Cyclin D1 is a downstream target of the Wnt pathway and is involved in regulation of cell cycle progression. Reverse transcriptase PCR analysis showed decreased levels of cyclin D1. RNA was isolated after 48 hours of treatment with Wnt3a or Wnt3a with niclosamide. Cyclin D1 expression levels were downregulated by the increased concentration of niclosamide. Cyclin D1 was downregulated to 30%±15% in Huh-6 cells (P<0.05, ) and to 44%±14% in Hep3B cells (P<0.05, ) at 1.0 μM. Cyclin D1 expression levels were upregulated by increased Wnt3a concentrations to 236%±76% with 6 ng/mL Wnt3a in Huh-6 cells (P<0.05, ) and 180%±20% with 20 ng/mL Wnt3a in Hep3B cells (P<0.05, ). Niclosamide was added to medium containing 20 ng/mL Wnt3a and cultured for 48 hours. Cyclin D1 expression levels were downregulated to 35%±10% in Huh-6 (P<0.05, ) and 50%±15% in Hep3B () with 1 μM niclosamide treatment. These data clearly demonstrate that Wnt3a upregulates cyclin D1 expression levels and that niclosamide inhibits Wnt3a activity.
Figure 3 Real-time quantitative polymerase chain reaction. The expression level of cyclin D1 was analyzed by real-time quantitative polymerase chain reaction with Huh-6 cells (A, C, and E) and Hep3B cells (B, D, and F). Wnt3a upregulated cyclin D1 expression (C and D). Niclosamide downregulated cyclin D1 expression with (E and F) or without (A and B) Wnt3a (2 ng/ml).
Abbreviations: W, Wnt3a; Ni, niclosamide.
The TOPflash reporter plasmid contains three copies of the T-cell factor binding site, which is one of the targets of the Wnt pathway.Citation16 The promoter activity of TCF decreased to 8%±3% in Huh-6 cells (P<0.05, ) and 13%±8% in Hep3B cells (P<0.05, ) with 1 μM niclosamide. The promoter activity of TCF increased to 260%±30% in Huh-6 cells (P<0.05, ) and 200%±35% in Hep3B cells (P<0.05, ) with 6 ng/mL Wnt3a. The TCF promoter activity decreased to 0 in Huh-6 cells (P<0.05, ) and to 20%±10% in Hep3B cells (P<0.05, ) with 1 μM niclosamide treatment. Expression levels of β-catenin, DVL2, and cyclin D1 were analyzed using Western blot analysis (). One μM niclosamide suppressed expression levels with or without 20 ng/mL Wnt3a.
Figure 4 Luciferase assay. A luciferase assay was performed with TOPflash after the addition of Wnt3a or niclosamide to Huh-6 cells (A, C, and E) and Hep3B cells (B, D, and F). Luciferase activity of TOPflash was calculated as relative light units divided by that of pRL-TK. Wnt3a increased the luciferase activity of TOPflash (C and D). Luciferase activity was suppressed by niclosamide with (E and F) or without (A and B) Wnt3a (2 ng/ml).
Abbreviations: W, Wnt3a; Ni, niclosamide.
Discussion
Niclosamide suppressed cell proliferation to 70% at 1 μM in the human colon cancer cell line HCT116, and at 1.5 μM in the human ovarian cancer cell line CP70.Citation14 In our experiments, 1 μM niclosamide suppressed cell proliferation to 29% in Huh-6 cells and 41% in Hep3B cells without Wnt3a, and to 17% in Huh-6 cells and 62% in Hep3B cells with Wnt3a. These results are comparable with those obtained in previous reports. We added niclosamide directly to hepatoma cells without serum or Wnt3a and found that niclosamide suppressed cell proliferation. Huh-6 cells expressed all Fz genes except Fz8, while Hep3B expressed all but Fz1 and Fz8.Citation6 This suggests that the Wnt pathway is constitutively active and inhibited by niclosamide. Sack et al suggested that niclosamide inhibits formation of the β-catenin/TCF complex.Citation11 Previous reports and our present data indicate that niclosamide may suppress cell proliferation with or without Wnt ligands. This hypothesis is supported by the fact that hepatoma cell proliferation is suppressed by sorafenib in serum-free culture.Citation17
Dysregulation of the Wnt pathway has been implicated in various types of cancer, including hepatoma.Citation18 Wnt3a is upregulated in human hepatoma tissues.Citation1 It is speculated that Wnt3a is involved in the growth of hepatoma cells and increases the promoter activity of TCF.Citation19 With this upregulation, TCF binds to the promoter of cyclin D1 and increases its expression.Citation20 Our study clearly demonstrates that Wnt3a increased the proliferation of Huh-6. Moreover, Wnt3a increased expression of cyclin D1 and stimulated the promoter activity of TCF with TOPflash. These data suggest that Wnt3a activates the Wnt pathway, upregulates cyclin D1 expression, and promotes proliferation of Huh-6 cells, consistent with previous reports.
The role of the Wnt pathway in hepatocarcinogenesis is intriguing. Hepatitis C core proteins are involved in hepatocarcinogenesisCitation21 and upregulation of Wnt3a and Fz1 in SMMC-7721 cells.Citation22 Interestingly, the promoter activity of TCF is stronger with the HCV core protein and Wnt3a than it is with Wnt3a alone.Citation19 Yang et al cultured HepG2 cells with Wnt3aCitation23 and found an upregulation of Kruppel-like factor 8, an oncogenic transcription factor leading to increased β-catenin accumulation. For the treatment of hepatoma, inhibition of the activated Wnt pathway is essential. In our present study, niclosamide successfully inhibited the Wnt pathway and suppressed proliferation of Huh-6 cells. Our data are in accordance with the previous report by Sack et al.Citation11
In this study, niclosamide suppressed the promoter activity of TCF. Our present data corroborate our previous report and suggest that niclosamide inhibits the Wnt pathway. Our data clearly show that niclosamide successfully suppresses hepatoma cell proliferation. shows that niclosamide induces apoptosis as evidenced by the presence of pyknotic nuclei.Citation24
Human hepatoma tissues harbor TUNEL-positive cells and proliferation cell nuclear antigen-positive cells.Citation25 Thus, the results of the cell proliferation assay should reflect the accumulative results of both effects. Niclosamide antagonized Wnt3a-enhanced cell proliferation () while inducing cell apoptosis. The accumulative result was, therefore, indicative of hepatoma, and our results showed a decrease in total cell number.
Niclosamide inhibits the Wnt signaling pathway by internalizing Fz1. However, Fz1 is not upregulated in cancerous tissues.Citation1 Use of niclosamide as a treatment of hepatoma, therefore, may have limitations when translating the results of this study to the clinic.
In conclusion, niclosamide suppressed the proliferation of hepatoma cells and may be a potential candidate agent for the treatment of hepatoma.
Acknowledgments
This work was supported in part by a Research Grant-in-Aid for Scientific Research (23591002) from the Japan Society for the Promotion of Science.
Disclosure
The authors report no conflicts of interest in this work.
References
- BengocheaAde SouzaMMLefrancoisLCommon dysregulation of Wnt/Frizzled receptor elements in human hepatocellular carcinomaBr J Cancer200899114315018577996
- Takahashi-YanagaFActivator or inhibitor? GSK-3 as a new drug targetBiochem Pharmacol201386219119923643839
- TanakaSSKojimaYYamaguchiYLNishinakamuraRTamPPImpact of WNT signaling on tissue lineage differentiation in the early mouse embryoDev Growth Differ201153784385621762130
- KatohMKatohMWNT signaling pathway and stem cell signaling networkClin Cancer Res200713144042404517634527
- HendrickxMLeynsLNon-conventional Frizzled ligands and Wnt receptorsDev Growth Differ200850422924318366384
- FujimotoTTomizawaMYokosukaOSiRNA of frizzled-9 suppresses proliferation and motility of hepatoma cellsInt J Oncol200935486186619724923
- KnorrRTreatment of tapeworm with Yomesan in 36 patientsMed Lav19605519371938 German15445386
- MwapeKEPhiriIKPraetNTaenia solium Infections in a rural area of Eastern Zambia-a community based studyPLoS Negl Trop Dis201263e159422479664
- BalgiADFonsecaBDDonohueEScreen for chemical modulators of autophagy reveals novel therapeutic inhibitors of mTORC1 signalingPLoS One200949e712419771169
- XieZNairUKlionskyDJAtg8 controls phagophore expansion during autophagosome formationMol Biol Cell20081983290329818508918
- SackUWaltherWScudieroDNovel effect of antihelminthic niclosamide on S100A4-mediated metastatic progression in colon cancerJ Natl Cancer Inst2011103131018103621685359
- ChenMWangJLuJThe anti-helminthic niclosamide inhibits Wnt/Frizzled1 signalingBiochemistry20094843102671027419772353
- OsadaTChenMYangXYAntihelminth compound niclosamide downregulates Wnt signaling and elicits antitumor responses in tumors with activating APC mutationsCancer Res201171124172418221531761
- YoYTLinYWWangYCGrowth inhibition of ovarian tumor-initiating cells by niclosamideMol Cancer Ther20121181703171222576131
- TomizawaMSaishoHInsulin-like growth factor (IGF)-II regulates CCAAT/enhancer binding protein alpha expression via phosphatidylinositol 3 kinase in human hepatoblastoma cell linesJ Cell Biochem2007102116117017372916
- Leung-HagesteijnCMahendraANaruszewiczIHanniganGEModulation of integrin signal transduction by ILKAP, a protein phosphatase 2C associating with the integrin-linked kinase, ILK1EMBO J20012092160217011331582
- TomizawaMShinozakiFSugiyamaTYamamotoSSueishiMYoshidaTSorafenib suppresses the cell cycle and induces the apoptosis of hepatocellular carcinoma cell lines in serum-free mediaExp Ther Med20101586386622993610
- ZenderLVillanuevaATovarVSiaDChiangDYLlovetJMCancer gene discovery in hepatocellular carcinomaJ Hepatol201052692192920385424
- LiuJDingXTangJEnhancement of canonical Wnt/beta-catenin signaling activity by HCV core protein promotes cell growth of hepatocellular carcinoma cellsPLoS One2011611e2749622110662
- Takahashi-YanagaFSasaguriTGSK-3beta regulates cyclin D1 expression: a new target for chemotherapyCell Signal200820458158918023328
- BanerjeeARayRBRayROncogenic potential of hepatitis C virus proteinsViruses2010292108213321994721
- LiuJWangZTangJHepatitis C virus core protein activates Wnt/beta-catenin signaling through multiple regulation of upstream molecules in the SMMC-7721 cell lineArch Virol201115661013102321340743
- YangTCaiSYZhangJKruppel-like factor 8 is a new Wnt/beta-catenin signaling target gene and regulator in hepatocellular carcinomaPLoS One201276e3966822761862
- TomizawaMShinozakiFSugiyamaTYamamotoSSueishiMYoshidaTInsulin-like growth factor-I receptor in proliferation and motility of pancreatic cancerWorld J Gastroenterol201016151854185820397262
- HinoNHigashiTNousoKNakatsukasaHTsujiTApoptosis and proliferation of human hepatocellular carcinomaLiver19961621231298740846