?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Aim: Human induced pluripotent stem cells (iPSCs) are inefficiently derived from somatic cells by overexpression of defined transcription factors. Overexpression of H2A histone variant macroH2A1.1, but not macroH2A1.2, leads to increased iPSC reprogramming by unclear mechanisms. Materials & methods: Cleavage under targets and tagmentation (CUT&Tag) allows robust epigenomic profiling of a low cell number. We performed an integrative CUT&Tag-RNA-Seq analysis of macroH2A1-dependent orchestration of iPSCs reprogramming using human endothelial cells. Results: We demonstrate wider genome occupancy, predicted transcription factors binding, and gene expression regulated by macroH2A1.1 during reprogramming, compared to macroH2A1.2. MacroH2A1.1, previously associated with neurodegenerative pathologies, specifically activated ectoderm/neural processes. Conclusion: CUT&Tag and RNA-Seq data integration is a powerful tool to investigate the epigenetic mechanisms occurring during cell reprogramming.
Cleavage under targets and tagmentation (CUT&Tag) is a methodology, devised in 2019, that is used for efficient epigenomic profiling of small samples and single cells [Citation1]. CUT&Tag-Seq brings together antibody-targeted controlled cleavage by protein A-Tn5 fusion with massive parallel DNA sequencing to discover the binding sites of histone post-translational modifications (PTMs), RNA polymerase II and transcription factors [Citation1–3] at the tissue scale and cellular level. CUT&Tag can capture epigenomic heterogeneity and allows prediction of sensitivity to therapeutic agents in samples taken from leukemic patients [Citation2]. While chromatin immuno-precipitation sequencing (ChIP-Seq) is the most common technique employed to analyze protein–DNA interactions, CUT&Tag offers several advantages, such as the fact that it does not require cell lysis or chromatin fractionation, and it is also ideal for single-cell platforms [Citation1]. CUT&Tag was recently combined with RNA-seq to simultaneously profile histone PTMs and gene expression in single cells isolated from mouse brain tissue [Citation4]. ‘Multiomics’ approaches consisting of the integrated analysis of transcriptional activity, histone modifications and chromatin accessibility using CUT&Tag are still underdeveloped compared with established ChIP-seq datasets, although they have the potential to uncover the intricate epigenetic regulatory mechanisms governing different cell types.
Induced pluripotent stem cells (iPSCs) are pluripotent stem cells that can be produced from a somatic cell by the introduction of four specific transcription factors (namely Myc, Oct3/4, Sox2 and Klf4) key to the process [Citation5]. iPSCs are used in personalized medicine modeling and approaches, and have great potential in autologous-based regenerative medicine [Citation5]. However, they are generally not considered safe when transplanted because of their inherent iatrogenic tumorigenesis [Citation6]. There are several potential causes for tumorigenicity during the induction of pluripotency in somatic cells, which is linked to genome and epigenome stability [Citation7]. The process of reprogramming involves a complete remodeling of the somatic epigenetic memory, which is replaced by new iPSC-specific epigenetic patterns. Among epigenetic alterations, the substitution of canonical histones with histone variants has recently emerged as a regulator of iPSC identity [Citation8].
MacroH2A proteins are histone variants coded by two distinct genes: H2AFY for macroH2A1 and H2AFY2 for macroH2A2. Unlike macroH2A2, macroH2A1 is expressed ubiquitously. MacroH2A1 features two exon-splicing isoforms – macroH2A1.1 and macroH2A1.2 – which have common and distinct biological functions in tumorigenesis, cell differentiation and stemness [Citation9–21]. In particular, it was recently reported that macroH2A1.1, but not macroH2A1.2, is an enhancer of DNA damage repair and iPSC reprogramming from somatic cells, increasing the efficiency of the process [Citation22]. Interestingly, while macroH2A1.2 does not affect iPSC differentiation into the three embryonic germ layers (endoderm, mesoderm and ectoderm), macroH2A1.1-expressing reprogrammed iPSCs have less potential to generate ectoderm [Citation22].
Understanding the genome-binding and transcriptomic dynamics of macroH2A1.1 could help to improve iPSC production and availability for clinical trials. However, iPSC reprogramming remains an inefficient process, and the amount of cells upon somatic reprogramming is unsuitable for conventional ChIP-Seq approaches. In this work, we employed human umbilical vein endothelial cells (HUVECs) during their reprogramming into iPSCs overexpressing tagged macroH2A1.1 or macroH2A1.2 to achieve for the first time an integrative CUT&Tag/RNA-Seq analysis of histone variant macroH2A1-dependent regulation of human iPSC reprogramming from somatic cells. Our work uncovers a novel histone variant-based genomic/transcriptomic interplay underlying iPSC reprogramming, which may inform functional and preclinical assays.
Materials & methods
Cell reprogramming
HUVECs were obtained from Thermo Fisher Scientific (MA, USA). Cells were cultured in Nunc EasYFlasks in Endothelial Cell Growth medium 2 (Promocell, Heidelberg, Germany). Cellular reprogramming was performed as described in [Citation22].
CUT&Tag, library preparation, & sequencing
Cells on the 4th day post-transfection were collected by TryPLE Express Enzyme (Thermo Fisher Scientific, Wien, Austria) in 1.5-ml tubes. CUT&Tag was performed according to the manufacturer’s protocol (Active Motif, CA, USA). Briefly, cells were harvested and washed once using 1 ml of 1× wash buffer. In parallel, the concanavalin A beads were prepared. Every 500,000 cells, 20 μl of the beads’ slurry were withdrawn and mixed with 1.6 ml of 1× binding buffer. Beads were harvested using a magnetic stand and washed twice with 1.5 ml of 1× binding buffer. The pellet containing previously harvested cells and the bead suspension were mixed in a 1.5-ml tube, rotating end-over-end for 10 min at room temperature. Cells were harvested using a magnetic stand to clear and the supernatant was discarded. The pellet was resuspended in 50 μl antibody buffer and 1 μl rabbit ab9108 anti-6× His tag antibody (Abcam, Cambridge, UK) was added where needed. The mix was incubated overnight at 4°C using orbital mixing to ensure the liquid stayed together at the bottom of the tube. The guinea pig anti-rabbit antibody secondary antibody was diluted 1:100 in dig-wash buffer. The tubes were cleared using a magnetic stand and the liquid was discarded. A total of 100 μl of guinea pig anti-rabbit antibody mix was added and tubes were incubated at room temperature for 60 min in an orbital rotator. Cells were cleared through a magnetic stand, and they were washed three times using 1 ml dig-wash buffer. In parallel, the CUT&TAGTM assembled pA-Tn5 transposomes were diluted 1:100 with Dig-300 buffer. For each immunoprecipitated sample, 100 μl of CUT&TAGTM assembled pA-Tn5 transposomes was added, and the mix was placed on an orbital rotator for 60 min at room temperature. The cells were harvested using a magnetic stand and eventually washed three times using 1 ml Dig-300 buffer. In the following step each tube was supplemented with 125 μl Tagmentation Buffer and incubated at 37°C for 60 min. To isolate the DNA surrounding macroH2A1.1 and macroH2A1.2 deposition regions, the tagmentation was first stopped by the addition of 4.2 μl of 0.5 M EDTA, 1.25 μl of 10% SDS and 1.25 μl of proteinase K (10 mg/ml) per each sample. The tubes were then vortexed at full speed and incubated for 60 min at 55°C to digest. The samples were cleared using a magnetic stand and 625 μl of DNA purification buffer per each was added. The samples were transferred to DNA purification columns and centrifuged at 17,000 × g for 1 min. After the flow-through was discarded, 750 μl of DNA purification wash buffer was added to each sample, which was then centrifuged again at 17,000 × g for 1 min. The flow-through was discarded. To remove any leftover DNA purification wash buffer, the empty columns were further centrifuged at 17,000 × g for 2 min. To isolate the DNA, 35 μl of DNA purification elution buffer was added to each sample, which was then incubated at room temperature for 1 min and eventually centrifuged at 17,000 × g for another 1 min.
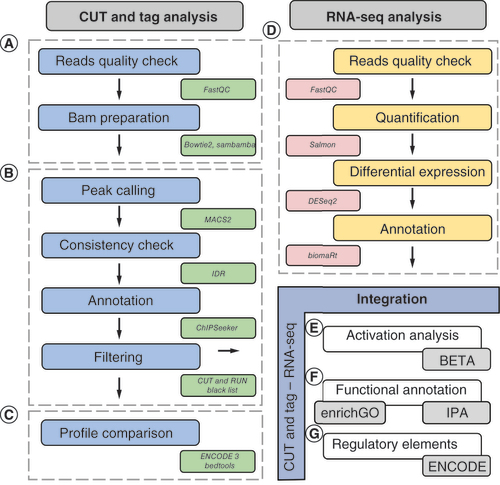
Library preparation for CUT&Tag samples was performed according to the manufacturer’s protocol (Active Motif). Paired-end Illumina sequencing was performed using an Illumina Hiseq 2500 (Illumina, CA, USA), obtaining two FASTQ files for each sample. MacroH2A1.1, macroH2A1.2 and the control samples were sequenced at least in duplicate, and a total of 4.8, 5.4 and 4.8 million reads were generated. These reads underwent integrative bioinformatics analysis, which is summarized in and described in the following sections.
Alignment & peak calling
Paired-end CUT&Tag FASTQ files were quality controlled using FastQC v0.11.9 [Citation23] and were aligned to the GRCh38 reference human genome using Bowtie2 v2.3.5.1 [Citation24] with the following parameters: -q –local –very-sensitive-local –no-mixed –no-discordant –phred33 -I 10 -X 700. The resulting BAM files were filtered to keep only uniquely mapped reads using the view module of sambamba v0.8.2 [Citation25] with the following parameters: -F ‘[XS] == null and not unmapped and not duplicate’ (A).
Peaks were called using MACS2 v2.2.6 [Citation26] for each histone variant’s biological replicate with the following blueprint command line: macs2 callpeak -t replicate_bam –broad -p 1e-5 -c control_bam -n sample_name -f BAMPE –keep-dup all –outdir out_dir, where we set the statistical significance threshold, the data format of the input bam file and the handling mode of duplicate tags with the options ‘-p’, ‘-f’ and ‘–keep-dup’, respectively. Prior to performing any downstream analyses on the named peaks, a quality check was performed using the ChiPQC v1.30.0 R package [Citation27]. To assess consistency between replicates, we resorted to the irreproducibility discovery rate (IDR), as implemented in the idr v2.0.4.2 R package [Citation28]. Peaks (Supplementary Figure 1) were annotated using the annotatePeak function of the ChIPSeeker v1.30.3 R package [Citation29], which consulted the TxDb.Hsapiens.UCSC.hg38.knownGene v3.10.0 UCSC table and generated an Entrez Gene ID whenever a ±1 kb window enclosing a gene’s transcription start site (TSS) overlapped with a peak. Peaks were discarded if included in the CUT&RUN Blacklist of Problematic Regions of the Genome (B) [Citation30].
Peaks exhibiting p-values ≤0.05 were retained for all downstream analyses involving integration with RNA-Seq data. When conducting in silico functional enrichment analysis of nearby genes prior to RNA-Seq data integration, peaks were selected more stringently by setting the significance threshold to 0.01.
Histone profiles comparison
Occupancy profiles of the two histone variants were compared with several public ENCODE 3 epigenomic datasets. In detail, 16 experiments matched the ‘query endothelial_cell_of_humbilical_vein’ as a biosample and contained histone ChIP-Seq data (Supplementary Table 1). All 16 experiments were individually compared with our study; using Bedtools v.2.26.0, we reported an intersection between their peaks and those found in this study whenever they shared at least 1 bp. The overall similarity was estimated using the Jaccard index:where A and B are the numbers of peaks for the datasets A and B in comparison, and
and
represent the dimensions of the intersection and union of these datasets, respectively (C).
RNA-Seq
RNA-Seq data were previously deposited into the Gene Expression Omnibus (GEO) repository with the identifier GSE164396. In our previous study [Citation22], we analyzed this dataset as follows: reads were quasi-mapped onto release 36 of the GENCODE human reference transcriptome and counted using Salmon v1.2.1 [Citation31] with standard parameters aside from the validateMappings flag, which was set to enable a selective alignment of the reads during mapping onto the transcriptome. Differential gene expression analysis was performed using DESeq2 v3.16 [Citation32] for both histone isoforms when compared with the control samples. DESeq2 was run with standard parameters. Genes exhibiting absolute log2 fold-change values ≥1.5 and Benjamini-Hochberg-adjusted p-values ≤0.05 were considered differentially expressed genes (DEGs) between contrasts (control samples vs MacroH2A1.1 and control samples vs macroH2A1.2). Genes were annotated using the biomaRt v3.17 R package (D). Therefore, RNA-Seq data were not reanalyzed, but we retrieved DEGs from [Citation22] and used them in this work for all the following analytical steps.
CUT&Tag & RNA-Seq data integration
The Entrez Gene IDs of genes near or including peaks were converted to Ensembl Gene IDs using the mygene v3.2.2 Python module, which were used, in turn, as cross-reference keys to match the DEGs. The resulting DEGs upon histone binding were referred to as UHB-DEGs. Relationships between high-quality peaks and RNA-Seq data were also analyzed through BETA v.1.0.7 [Citation33], which was run with the following blueprint command line: BETA basic -p peaks.bed -e gene_expr.bsf -k BSF -g hg38 -o out1 –gname2 –n macroH2A1.1 –da 500 –cutoff 1, where ‘BSF’ is a three-column file format containing the gene IDs, log2 fold-change expression values, and p-values; ‘–o’ and ‘–n’ define the output folder and file prefix; ‘–gname2’ indicates the type of gene IDs that in our case is the official gene symbol; ‘–da’ sets the number of top significantly differentially expressed genes to pick from the total pool; ‘–cutoff’ ranges from 0 to 1 and indicates the p-value cutoff for the BETA’s activation/repression prediction test. Thus, BETA was used to predict the global ‘activation’ or ‘repression’ role of macroH2A1.1 and 1.2 on gene expression, and calculate a potential regulatory score or rank product for genes proximal to peaks and predict them as potentially ‘activated’ or ‘repressed’. We have highlighted the genes resulting from the BETA analysis that exhibited a rank product ≤0.001 (suggested cut-off), absolute log2 fold-change values ≥1.5 and are UHB-DEGs in E.
Gene set & pathway-enrichment analyses
Functional annotation of CUT&Tag genes alone and integrated with DEGs was performed using the clusterProfiler v4.2.2 [Citation34] R packages and by resorting to the NCBI RefSeq functional elements, NCBI transcription factor-binding sites and NCBI DNAse hypersensitive sites UCSC datasets. Functional enrichment analysis against the biological process subset of the gene ontology (GO) was conducted using the clusterProfiler’s enrichGO function [Citation34,Citation35]. GO terms were considered significantly overrepresented if their q-values were ≤0.05 and then were summarized by REVIGO [Citation36]. Over-represented biological pathways were determined through the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Ingenuity Pathway Analysis (IPA); the latter was also used to infer pathway-activation states. In fact, IPA is based on the calculation of the activation z-score as a way to infer the activation states of predicted transcriptional regulators and pathways. Positive and negative z-scores are associated with activated and inhibited pathways, respectively. The more extreme a z-score, the more statistically confident a prediction is [Citation37]. Overrepresented pathways were considered significant if their p-values were ≤0.05 (F).
All results were plotted using the ggplot2 (v.3.3.5), enrichPlot (v.1.14.2) and circlize (v.0.4.15) R packages.
Histone binding site annotation
Binding sites near DEGs were evaluated for the presence of known regulatory elements. The genomic coordinates of all the elements reported in the encRegTfbsClustered dataset, a compilation of Transcription Factor ChIP-seq Clusters relative to 340 factors and 129 cell types from ENCODE 3 (ENCODE 3 - encRegTfbsClustered), were intersected with the peaks’ chromosomal coordinates by considering a minimum of 1 bp overlap and distance from the TSS included in the range -10–10 kbp. Overlapping regulatory sites obtained when searching for the ‘endothelial_cell_of_umbilical_vein’ keyword were retained if their score was ≥500; this score is calculated at UCSC, ranges from 1 to 1000 and is based on signal values assigned by the ENCODE pipeline (G).
Statistical analysis
The module chi2_contingency of the scipy.stats v1.7.3 Python package was used to test the differential distribution of peak frequency between chromosomes. A one-way χ2 test was performed using the chisquare module of scipy.stats. The tests were considered significant when p ≤0.05. A stacked plot of chromosomes was drawn by considering the normalized peak frequencies NPF, calculated as follows:
Results
Genome occupancy patterns of overexpressed macroH2A1 isoforms in HUVECs reprogrammed to iPSCs
We analyzed the genome occupancy patterns of 6-His-tagged macroH2A1.1 and macroH2A1.2 in HUVECs on the 4th day of their episomal-driven reprogramming into iPSCs [Citation22] by using a CUT&Tag approach. The 4th day of reprogramming was chosen for analysis because it corresponded to the peak of transient expression of either histone in HUVECs, preceding the full reprogramming of cells, occurring starting from 12 days post-transfection with the reprogramming episomes, leading in turn to the formation of alkaline phosphatase-positive (AP+) colonies and to the ability of iPSCs to differentiate into mesoderm, endoderm and ectoderm [Citation22]. A macroH2A1.2 replicate was dropped immediately after quality-control analysis (Supplementary Figure 2A). The identified broad peaks in the remaining replicates showed significant intragroup variability in terms of number and localization around the genes’ TSS of the detected peaks, presumably due to the inefficiency and variability of the iPSC reprogramming process (Supplementary Figure 2C). As internal positive controls, we found that both macroH2A1.1 and macroH2A1.2 exhibited peaks, before IDR, within LAMA5 e PTPRN gene bodies (Supplementary Figure 2C), as it is found consistently in macroH2A1 isoform ChIP-Seq datasets [Citation10]. Peaks exhibiting a significant p-value, that is, ≤0.05, as calculated by MACS2, were first subjected to IDR in order to keep only consistent peaks between replicates and then filtered using the CUT&RUN Blacklist of Problematic Regions of the Genome. Surviving peaks were 10,830 and 8872 for macroH2A1.1 and macroH2A1.2, respectively. Gene-wise annotation allowed us to count 7147 and 6280 peaks in the proximity of genes for macroH2A1.1 and macroH2A1.2, respectively, of which 3488 were shared between the two histone variants and 3659 and 2792 were unique (Supplementary Table 2 and Supplementary Table 3). The median peak sizes were 26.938 and 24.959 bp for macroH2A1.1 and macroH2A1.2 (Supplementary Figure 3A). Their genome-occupancy patterns and the distance from the TSS are plotted in the Supplementary Figure 4 left and right panels, respectively. MacroH2A1.1 and macroH2A1.2 peaks were found in promoters, distal intergenic regions and introns, with macroH2A1.2 showing a slight, not statistically significant preference for promoters (χ2[6, n = 19,702] = 4.93; p = 0.55) compared with macroH2A1.1. The majority of peaks in both isoforms were either near (1 kbp) or far away (>10 kbp) from TSS.
Comparison of occupancy patterns across histones
Genomic occupancy of macroH2A1.1 and macroH2A1.2 histone variants (10,830 and 8872 genomic regions, respectively) was compared with that of 16 different experiments for 12 different histones. Profiles were mostly different among experiments and histones, with low Jaccard index values; Supplementary Table 1, sheet S3) for most pairwise comparisons (mean J: 0.08; range: 10-5–0.47). Profiles were moderately overlapping among experiments targeting the H3K4me1/2/3 variants (mean J: 0.32), according to previous reports [Citation38]. MacroH2A1.1 and 1.2 resulted in a limited overlap (J: ∼0.05) with H2AFZ and H3K27ac profiles.
Gene-set & pathway-enrichment analyses of CUT&Tag data
Functional analysis performed directly on 128 and 543 genes located in the proximity of 129 macroH2A1.1 and 586 macroH2A1.2 peaks revealed that both macroH2A isoforms mainly enriched GO biological processes related to cell morphogenesis/development and the neuronal system. In particular, macroH2A1.1 displayed a more detailed enrichment of GO terms related to synaptic signaling, development and modulation (Supplementary Figure 5A) with fewer genes than macroH2A1.2, which in turn over-represented additional processes concerning cardiac activity and transmembrane transport/potential regulation (Supplementary Figure 5B). This indicates macroH2A1.1 as a more specific modulator of neuron-specific developmental processes compared with macroH2A1.2
Integration of CUT&Tag & transcriptomic data in HUVEC-overexpressing macroH2A1 isoforms reprogrammed to iPSCs
Peaks were further filtered according to whether they fell within the proximity of differentially expressed genes. Therefore, the surviving peaks were 231 for macroH2A1.1 and 46 for macroH2A1.2. The median peak sizes were 28,162 and 28,502 bp for macroH2A1.1 and macroH2A1.2 (Supplementary Figure 3B). We counted 131 UHB-DEGs for macroH2A1.1, of which 51 were significantly upregulated and 80 were downregulated, and 28 UHB-DEGs for macroH2A1.2, of which 15 were upregulated and 13 downregulated (A & Supplementary Table 4). Three UHB-DEGs, that is, PPP1R37, BACH2 and MEGF8, were shared between the two macroH2A1 variants. The genes PPP1R37 and BACH2 were upregulated, while MEGF8 was downregulated by both macroH2A1.1 and macroH2A1.2 overexpression in HUVEC cells. BETA analysis inferred 328 and 276 genes with regulatory scores <0.001 for macroH2A1.1 and 1.2, respectively, with a maximum genomic distance from peaks of 100 kbp (Supplementary Table 5, sheets ST1 & ST2). Among them, we identified 14 putatively repressed and 17 activated UHB-DEGs for macroH2A1.1 and ten repressed and nine activated for macroH2A1.2 (Supplementary Table 5, ST3 & ST4). Globally, macroH2A1.1 resulted in a very mild, although significant, repression activity (asymptotic two-sample Kolmogorov-Smirnov test, D = 0.09; p = 0.02) on transcripts, while no significant prediction was obtained for the other macroH2A variant.
(A) Volcano plots of the significant differentially expressed genes upon histone binding for macroH2A1.1 (left) and macroH2A1.2 (right). (B) Distribution of peaks in differentially expressed genes relative to the (left) gene structure and (right) distance from the nearest TSS.
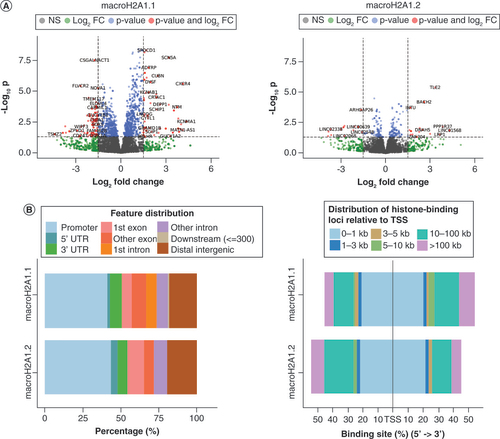
Peak occupancy patterns in UHB-DEGs were notably different from those preintegration with RNA-Seq data (Supplementary Figure 4). MacroH2A1.2 peaks were more frequent in promoters, 5′UTR and the first exon than macroH2A1.1 peaks were. On the contrary, peaks were more numerous in the first intron, other exons and 3′UTR of macroH2A1.1 (B, left). The peaks of macroH2A1.2 were absent between 3 and 5 Kbp upstream of the TSS and 5 and 10 Kbp downstream of the TSS (B, right).
UHB-DEG-related peak localization within chromosomes
After CUT&Tag and RNA-Seq data integration, no peaks were found in sexual chromosomes or in autosomes 18 and 21 for either isoform. MacroH2A1.1 peaks were present in all the remaining chromosomes apart from chromosome 16, where macroH2A1.2 peaks were instead abundant. In addition, macroH2A1.2 peaks were not found on chromosomes 2, 9 and 22, where macroH2A1.1 peaks were instead numerous (). The total number of peaks identified in chromosomes 2 (χ2[19, n = 277] = 4.77; p = 2.88e-02), 16 (χ2[19, n = 277] = 25.11; p = 5.42e-07), and 19 (χ2[19, n = 277] = 6.44; p = 1.11e-02) were statistically significantly different between the two isoforms in HUVECs undergoing reprogramming.
Bars are split into gene regions.
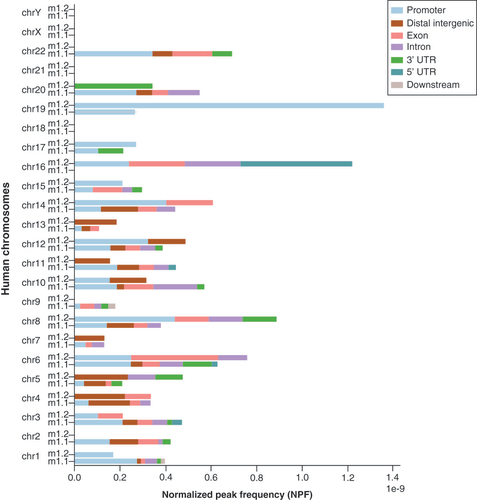
Considering gene structure, peak counts were not statistically different for any chromosome. Moreover, multiple peaks were found in the body or in proximity to a few UHB-DEGs for both isoforms. Eight peaks were found in the CSMD3 gene, seven in EPHB1, six in NRXN3 and NTM, and five in CRTAC1 for macroH2A1.1 but not for macroH2A1.2. The maximum number of peaks found in macroH2A1.2′s UHB-DEGs were, instead, five in two genes, that is, LINC01568 and BACH2, and three peaks in five genes (ZNF704, MATN2 and LINC00639; Supplementary Tables 6 & 7 for macroH2A1.1 and macroH2A1.2).
MacroH2A1.1 genome binding orchestrates the expression of more genes involved in functions related to the three different germ layers than macroH2A1.2
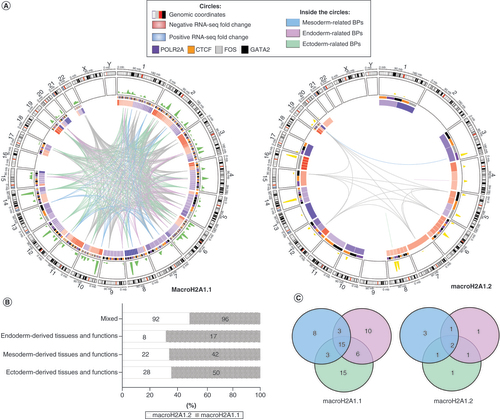
iPSCs are defined by their potential to differentiate into the three embryonic germ layers (ectoderm, mesoderm and endoderm). To follow-up the previous enrichment analysis of the genes associated with CUT&Tag peaks, we have conducted a further GO enrichment analysis of the UHB-DEGs. As macroH2A1.1, and not macroH2A1.2, was specifically associated with neural processes, and the nervous system is derived from the embryonic ectoderm, we manually categorized the enriched GO terms (Supplementary Tables 8 <& 9 for macroH2A1.1 and macroH2A1.2, respectively) based on the literature and their involvement in one of the three germ layers, that is, ectoderm, mesoderm and endoderm, or in more than one layer because of their multiple functions. This latter set of GO terms was named ‘mixed’. With a few exceptions, macroH2A1.2 UHB-DEGs were not specific to a single germ layer, while macroH2A1.1 UHB-DEGs could be associated more specifically with one of the three germ layers (A). Indeed, macroH2A1.1 peaks hit a significantly higher number of DEGs involved in biological processes related to the three different germ layers compared with macroH2A1.2. Frequencies of GO terms were comparable for the ‘mixed’ category of both isoforms. On the contrary, the number of germ layer-specific GO terms were systematically halved in macroH2A1.2 in respect to macroH2A1.1 (B). Focusing on the distinct genes making the three germ layer-specific GO terms, we found that they were more abundant in macroH2A1.1 with a ratio of 15:1 (ectoderm), 10:1 (endoderm) and ∼3:1 (mesoderm; C).
(A) Circos plot representing differentially expressed genes upon histone binding (UHB-DEGs) for macroH2A1.1 and macroH2A1.2, and several features related to their expression, gene ontology (GO) terms and CUT<&Tag peaks. From the outer to the inner tracks of the Circos plots: chromosomes, CUT<&Tag peaks, heatmap of fold-change expression values (red and blue mean upregulated and downregulated genes in macroH2A1.1 or macroH2A1.2 overexpressing cells), UHB-DEGs whose biological processes relate to mesoderm (light blue links), endoderm (light purple) and ectoderm (light green). Gray links were genes associated with more than one germ layer. (B) 100% stacked-bars of the frequencies of GO terms grouped by germ layers. (C) Venn diagram of UHB-DEGs associated with the three germ layers.
MacroH2A1 isoform-dependent signaling pathways regulating the stemness of iPSCs
KEGG was used as a starting point to further study the enrichment of macroH2A1.1 and macroH2A1.2 UHB-DEGs for biological pathways. MacroH2A1.1 turned out to be enriched in a few pathways, including axon guidance (hsa04360) and focal adhesion (hsa04510; Supplementary Table 10). These pathways are known to be involved in several biological mechanisms regulating the pluripotency of stem cells (hsa04550), such as MAPK (hsa04010), WNT (hsa04310), PI3K (hsa04151) and regulation of the actin cytoskeleton (hsa04810). In contrast, macroH2A1.2 exhibited a significant enrichment in pathways that do not contribute to the regulation of stemness in iPSCs (Supplementary Table 11). UHB-DEGs were additionally enriched using IPA, which is based on an orthogonal semantic knowledgebase and offers an estimate of the activation state of pathways. This was done as a cross-check for the previous analytical step. For macroH2A1.1, the total number of significant canonical pathways was 24 (Supplementary Table 12). Of these, several were neural development-related pathways, and one of them was predicted to be activated (semaphorin neuronal repulsive signaling pathway, z-score = 0.447). As a confirmation of KEGG analysis, IPA issued only one significantly enriched pathway, which was neither related to the nervous system nor predicted to be activated (Supplementary Table 13).
Regulatory signals in macroH2A1.1 & macroH2A1.2 UHB-DEGs
To gain insight into the transcriptional regulatory signals that could be mediated by macroH2A1.1 and macroH2A1.2 upon HUVEC reprogramming into iPCSs, we evaluated the isoforms’ capacity to mask/unmask transcription factor binding sites. This was accomplished by first querying for HUVEC-specific DNA regulatory features available from ENCODE 3′s EncRegTfbsClustered dataset. The available features for this cell line exclusively regarded the following DNA-binding proteins: CTCF, FOS, GATA2 and POLR2A. Among these features, 231 and 37 overlapped 82 and 16 histone occupancy regions, being localized near DEGs for macroH2A1.1 and macroH2A1.2, respectively, and mainly in promoters (for a global view, refer to A). MacroH2A1.1 and macroH2A1.2 binding regions included many CTCF-responsive elements, respectively near 51 and 11 DEGs (Supplementary Table 14). MacroH2A1.1 histone also overlapped with FOS and GATA2 binding sites near 41 and 13 DEGs, respectively; macroH2A1.2 turned out to likely influence FOS/GATA2-dependent regulation for seven and 1=one DEGs. Furthermore, the isoform binding sites intersected RNA polymerase (POL2RA) at its promoter regions, thereby directly controlling the expression of 16 and three genes for macroH2A1.1 and macroH2A1.2 (Supplementary Table 14). The expression of some genes, that is, PAK4, DEPP1, CUBN, CRTAC1 and LINC00607, was controlled by multiple bindings of macroH2A1.1 to the promoter regions of almost all CTCF, FOS, GATA2 and POL2RA DNA-binding proteins. Similarly, macroH2A1.2 appeared to regulate the expression of PPP1R37, BACH2, MATN2, LINC00639 and LINC02608 genes upon binding to GATA2, FOS and CTCF (Supplemental Table 14).
Discussion
It was previously reported that macroH2A histone variants (macroH2A1 and macroH2A2) indiscriminately and cooperatively function as an obstacle upon reprogramming towards pluripotency [Citation39]. The latter study further demonstrated that macroH2A2 is the main barrier to reprogramming. It was also recently reported that, in somatic cells, overexpression of the macroH2A1 splicing isoform macroH2A1.1-activated (but not macroH2A1.2-activated) transcriptional programs that instead improved iPSC reprogramming [Citation22]. By doing so, macroH2A1.1 overexpression enhanced iPSC reprogramming, suggesting that this splicing isoform could be a promising epigenetic target to ameliorate iPSC genome stability and therapeutic potential [Citation22]. Considering the formation of AP+ colonies as a readout, while we observed an increase of approximately 40% in iPSC formation in HUVEC-overexpressing macroH2A1.1 [Citation22], Gaspar-Maia et al. observed an increase of approximately 170–240% in iPSC formation in fibroblasts double knocked out for macroH2A1 (hence for both macroH2A1.1 and macroH2A1.2) and macroH2A2 [Citation39]. The experimental conditions between our work and Gaspar-Maia et al. were different in terms of:
Starting material (human cells[HUVECs] vs mouse cells [fibroblasts]);
Transient overexpression of macroH2A1.1 (hence only one isoform of macroH2A1, separate from macroH2A1.2) reaching a peak of approximately 80% of transfection efficiency on the 4th day post-transfection to then decrease sharply after the 8th day post-transfection, with analysis of AP+ colonies formation on the 12th day post-transfection [Citation22] versus constitutive genetic depletion of macroH2A1.1/macroH2A1.2/macroH2A2 [Citation39];
Method of reprogramming: we used episomal vectors containing five reprogramming factors (Oct4, Sox2, Lin28, Klf4 and Myc) [Citation22], while Gaspar-Maia et al. used a polycystronic lentiviral vector containing four reprogramming factors (Oct4, Sox2, Klf4 and Myc) for iPSC reprogramming [Citation39].
Even if the studies are not directly comparable, altogether the evidence suggest that macroH2A2 and macroH2A1.2, but not macroH2A1.1, could be members of the macroH2A family responsible for the inhibition of iPSC reprogramming [Citation22,Citation39].
Here, we shed further light on this process: using HUVEC cells undergoing episomal reprogramming into iPSCs, we provide an integrated analysis of macroH2A1.1 and macroH2A1.2 genome occupancy and transcriptional effects using CUT<&Tag and RNA-Seq, respectively. Unfortunately, it was not possible to determine endogenous protein expression levels in iPSCs due to the very low number of cells derived from immunoblotting. As previously described [Citation22], overexpression of macroH2A1 isoforms was monitored in HUVECs undergoing reprogramming using the 6-His-Tag in signal super-resolution confocal microscopy [Citation22]. The same antibodies against the 6-His-Tag were used both in the past study [Citation22] and in the CUT<&Tag approaches of the current manuscript, which could identify cells specifically overexpressing macroH2A1.1 or macroH2A1.2. The roles of macroH2A1 isoforms in regulating gene expression are apparently contradictory, and the underlying mechanisms are not well understood. Until recently, macroH2A1 was generally believed to play a role in transcriptional repression. However, in several instances its genome occupancy also correlates with active transcription of a subset of genes [Citation17,Citation38,Citation40], as we have shown here and in our previous study [Citation22]. Our data clearly demonstrate that on the 4th day of reprogramming, macroH2A1.2 DNA binding sites are more frequent in promoters and closer to the TSS than macroH2A1.1′s are in DEGs, while macroH2A1.1 binding sites are more prevalent in the 3′UTR and in introns. These genomic binding patterns are remarkably similar to those reported in liver cancer cells overexpressing macroH2A1.1 and macroH2A1.2 coupled to green fluorescent protein using ChIP-Seq [Citation41]. The consistent differences in their genomic distribution might be reflected in the limited overlap between the genes deregulated by the different macroH2A1 isoforms in physiological and pathological processes [Citation10]. In breast cancer cells, macroH2A1.1 can have either an inhibitory or a stimulating effect on target gene transcription by RNA polymerase II, depending on the chromatin landscape and on differential recruitment [Citation42]. Overall, macroH2A1.1 deposition on DEGs was more abundant, and chromosome mapping revealed more prevalent coverage compared with macroH2A1.2 in HUVECs reprogrammed to iPSCs. Interestingly, none of the macroH2A1 isoforms were present on the X chromosome of our male HUVECs (XY), which is consistent with its deposition on the inactive X chromosome in female cells, as macroH2A1 was originally identified [Citation43–45].
The molecular divergence between the two splice isoforms, macroH2A1.1 and macroH2A1.2, comprises the usage of a mutually exclusive exon containing a number of amino acids that define the shape and hydrophobicity of a pocket in the macrodomain [Citation46]. As a result, macroH2A1.1, but not macroH2A1.2, is able to bind NAD+-derived ADP-ribose. Adequate activation of NAD+-dependent pathways is crucial for iPSC pluripotency, conferring a metabolic advantage and enhancing resistance to cellular stress through kinase inhibition [Citation47–49]. A tempting hypothesis is that enhanced sensing of NAD+ signaling mediated by macroH2A1.1 may reflect an increased efficiency of iPSC reprogramming [Citation22]. During myogenic differentiation, the macroH2A1.1 metabolite-binding macrodomain was indispensable to support optimal mitochondrial function but not required for gene regulation [Citation50]; it remains to be elucidated whether NAD+-derived ADP-ribose binding would alter macroH2A1.1 genome occupancy in iPSCs undergoing reprogramming.
Moreover, the macroH2A1.1 binding sites hit a significantly higher number of DEGs involved in biological pathways related to the three germ layers (ectoderm, endoderm and mesoderm) than macroH2A1.2, generating more interconnected networks; all predicted macroH2A1.1 activating pathways were related to neuronal pathways. We have previously shown that macroH2A1.1-overexpressing reprogrammed iPSCs are less able to generate the ectoderm [Citation22]. Interestingly, macroH2A1 levels are upregulated in the tissues of patients suffering from Huntington’s disease [Citation51] and Alzheimer’s disease [Citation52]. In Huntington’s and Alzheimer’s disease, human embryonic stem cells display ectodermal anomalies during development [Citation53,Citation54]. Moreover, systemic depletion of macroH2A1.1 in mice supports an epigenetic control necessary for increased hippocampal function and social behavior [Citation19]. Harnessing macroH2A1.1 in iPSC-derived neural cells may offer useful tools for studying neurological disease molecular mechanisms and developing therapies [Citation55]. Our findings also show that, in HUVEC cells, macroH2A1.1 binding sites overlapped with binding sites for transcription factors CTCF, FOS, GATA2 and POLR2A to a greater extent than macroH2A1.2. These transcription factors have been consistently involved in embryonic development and in the regulation of ectoderm specification towards the nervous system in several organisms [Citation56–59], as well as in iPSC reprogramming [Citation60–63].
Using a pan-macroH2A1 isoform ChIP-antibody (not discriminating between the two isoforms), Gaspar-Maia et al. reported that target genes (i.e., transcription factors Sall1 and Sall4) of the histone demethylase Utx, a critical enzyme in the induction of iPSC reprogramming and required for proper induction of ectoderm and mesoderm during differentiation of embryonic stem cells, are enriched for macroH2A in fibroblasts reprogrammed into iPSCs [Citation39]. Our CUT<&Tag data set profiles are not consistent with these findings, possibly due to the divergent species and cell types of origin considered, as mentioned earlier. Moreover, although we consistently found regulation of the mRNA levels of genes required to maintain or achieve pluripotency (Myc, Oct4, Sox2 and Klf4), upon overexpression of macroH2A1.1 in HUVEC cells [Citation22] our CUT<&Tag analysis did not identify significant peaks confirming their direct genome occupancy by macroH2A1.1, as shown by Gaspar-Maia et al. [Citation39]. Our findings suggest that macroH2A1.1 orchestrates pluripotency in human iPSCs via indirect transcriptional regulation of pluripotency genes. A limitation of our transcription factor analysis is that it is restricted to HUVEC-specific DNA regulatory features present in the ENCODE 3′s EncRegTfbsClustered dataset. Moreover, CUT<&Tag is a new technique that has not been extensively benchmarked against existing ChIP-Seq datasets on the same biological samples. A comprehensive benchmarking of CUT<&Tag for H3K27ac and H3K27me3 against published ChIP-Seq profiles from ENCODE in K562 cells identified an approximate 50% recovery of known ENCODE peaks. However, the peaks identified by CUT<&Tag represent the strongest ENCODE peaks, with identical functional and biological enrichments as ChIP-seq peaks [Citation64]. Moreover, the iPSC reprogramming process is notoriously inefficient and variable. This has been highlighted by numerous publications [Citation65–68]. The heterogeneity in reprogramming occurring on the 4th day could be likely due to different proportions of differentiated cell types within each experiment, despite following the same protocol with the same iPSC lines. In line with this, our experiment revealed a significant variability among replicates, which required us to apply conservative filtration strategies to keep only highly informative peaks.
Conclusion
We propose that multi-omics CUT&Tag and RNA-Seq complementary data integration is a powerful, streamlined and cost-effective alternative tool to elucidate epigenetic and gene regulatory mechanisms occurring during cell reprogramming and lineage commitment.
MacroH2A1 histone variant exon-spliced isoforms (macroH2A1.1 and macroH2A1.2) are emerging regulators of induced pluripotent stem cell identity.
CUT&Tag allows robust macroH2A1-dependent epigenetic profiling of human umbilical vein endothelial cell-derived induced pluripotent stem cells.
Integrated analysis of CUT&Tag/RNA-Seq uncovered macroH2A1.1-dependent activation of ectoderm/neural-dependent pathways.
Conceptualization: T Mazza and M Vinciguerra. Methodology: N Liorni, A Napoli and S Castellana. Software: N Liorni. Formal analysis: N Liorni, A Napoli, S Castellana, S Giallongo and T Mazza. Investigation: S Giallongo, O Lo Re and D Řeháková. Resources: D Řeháková and I Koutná. Data curation: N Liorni, S Giallongo and O Lo Re. Writing – original draft: N Liorni, T Mazza and M Vinciguerra. Writing – review and editing: N Liorni, A Napoli, S Castellana, DR, O Lo Re, I Koutná, T Mazza and M Vinciguerra. Supervision: I Koutná, T Mazza and M Vinciguerra. Project administration: M Vinciguerra. Funding acquisition: T Mazza and M Vinciguerra.
Competing interests disclosure
The authors have no competing interests or relevant affiliations with any organization or entity with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Writing disclosure
No writing assistance was utilized in the production of this manuscript.
Supplemental Information
Download Zip (4 MB)Acknowledgments
The authors thank Genomix4Life (Baronissi, Italy) and CEITEC (Brno, Czech Republic) for excellent technical assistance with genomics and transcriptomics approaches, respectively.
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: www.tandfonline.com/doi/suppl/10.2217/epi-2023-0267
Financial disclosure
This research was funded by the European Regional Development Fund-Project MAGNET (CZ.02.1.01/0.0/0.0/15_003/0000492), the Ministry of Health of the Czech Republic (NV18-03-00058), the Italian Ministry of Health and ‘5x1000’ voluntary contributions (R22-5x1000), and by the European Commission Horizon 2020 Framework Program (project 856871—TRANSTEM). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Data sharing statement
The RNA-seq data [Citation22] was deposited into GEO with the dataset identifier GSE164396. Genomics sequences obtained in this study by CUT&Tag has been deposited into GEO with the dataset identifier GSE214013.
References
- Kaya-Okur HS , WuSJ , CodomoCAet al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun.10, 1930 (2019).
- Janssens DH , MeersMP , WuSJet al. Automated CUT&Tag profiling of chromatin heterogeneity in mixed-lineage leukemia. Nat. Genet.53, 1586–1596 (2021).
- Bartosovic M , KabbeM , Castelo-BrancoG. Single-cell CUT&Tag profiles histone modifications and transcription factors in complex tissues. Nat. Biotechnol.39, 825–835 (2021).
- Zhu C , ZhangY , LiYE , LuceroJ , BehrensMM , RenB. Joint profiling of histone modifications and transcriptome in single cells from mouse brain. Nat. Methods18, 283–292 (2021).
- Takahashi K , YamanakaS. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell126, 663–676 (2006).
- Lee AS , TangC , RaoMS , WeissmanIL , WuJC. Tumorigenicity as a clinical hurdle for pluripotent stem cell therapies. Nat. Med.19, 998–1004 (2013).
- Liang G , ZhangY. Genetic and epigenetic variations in iPSCs: potential causes and implications for application. Cell Stem Cell13, 149–159 (2013).
- Giallongo S , RehakovaD , RaffaeleM , LoRe O , KoutnaI , VinciguerraM. Redox and epigenetics in human pluripotent stem cells differentiation. Antioxid. Redox Signal.34, 335–349 (2021).
- Bereshchenko O , LoRe O , NikulenkovFet al. Deficiency and haploinsufficiency of histone macroH2A1.1 in mice recapitulate hematopoietic defects of human myelodysplastic syndrome. Clin. Epigenetics11, 121 (2019).
- Borghesan M , FusilliC , RappaFet al. DNA hypomethylation and histone variant macroH2A1 synergistically attenuate chemotherapy-induced senescence to promote hepatocellular carcinoma progression. Cancer Res.76, 594–606 (2016).
- Giallongo S , DiRosa M , CaltabianoRet al. Loss of macroH2A1 decreases mitochondrial metabolism and reduces the aggressiveness of uveal melanoma cells. Aging12, 9745–9760 (2020).
- Giallongo S , LoRe O , LochmanováGet al. Phosphorylation within Intrinsic disordered region discriminates histone variant macroH2A1 splicing isoforms-macroH2A1.1 and macroH2A1.2. Biology10, doi: 10.3390/biology10070659 (2021).
- Lo Re O , DouetJ , BuschbeckMet al. Histone variant macroH2A1 rewires carbohydrate and lipid metabolism of hepatocellular carcinoma cells towards cancer stem cells. Epigenetics13, 829–845 (2018).
- Lo Re O , MazzaT , GiallongoSet al. Loss of histone macroH2A1 in hepatocellular carcinoma cells promotes paracrine-mediated chemoresistance and CD4+CD25+FoxP3+ regulatory T cells activation. Theranostics10, 910–924 (2020).
- Pazienza V , BorghesanM , MazzaTet al. SIRT1-metabolite binding histone macroH2A1.1 protects hepatocytes against lipid accumulation. Aging6, 35–47 (2014).
- Pazienza V , PanebiancoC , RappaFet al. Histone macroH2A1.2 promotes metabolic health and leanness by inhibiting adipogenesis. Epigenetics Chromatin9, 45 (2016).
- Podrini C , KoffasA , ChokshiSet al. MacroH2A1 isoforms are associated with epigenetic markers for activation of lipogenic genes in fat-induced steatosis. FASEB J.29, 1676–1687 (2015).
- Rappa F , GrecoA , PodriniCet al. Immunopositivity for histone macroH2A1 isoforms marks steatosis-associated hepatocellular carcinoma. PLOS ONE8, e54458 (2013).
- Chiodi V , DomeniciMR , BiaginiTet al. Systemic depletion of histone macroH2A1.1 boosts hippocampal synaptic plasticity and social behavior in mice. FASEB J.35, e21793 (2021).
- Guberovic I , FarkasM , CorujoD , BuschbeckM. Evolution, structure and function of divergent macroH2A1 splice isoforms. Semin. Cell Dev. Biol.135, 43–49 (2023).
- Lo Re O , VinciguerraM. Histone MacroH2A1: a chromatin point of intersection between fasting, senescence and cellular regeneration. Genes8, doi: 10.3390/genes8120367 (2017).
- Giallongo S , ŘehákováD , BiaginiTet al. Histonevariant macroH2A1.1 enhances nonhomologous end joining-dependent DNA double-strand-break repair and reprogramming efficiency of human iPSCs. Stem Cells40, 35–48 (2022).
- Andrews S et al. FastQC: a quality control tool for high throughput sequence data (2010).
- Langmead B , SalzbergSL. Fast gapped-read alignment with Bowtie 2. Nat. Methods9, 357–359 (2012).
- Tarasov A , VilellaAJ , CuppenE , NijmanIJ , PrinsP. Sambamba: fast processing of NGS alignment formats. Bioinformatics31, 2032–2034 (2015).
- Zhang Y , LiuT , MeyerCAet al. Model-based analysis of ChIP-Seq (MACS). Genome Biol.9, R137 (2008).
- Carroll TS , LiangZ , SalamaR , StarkR , de SantiagoI. Impact of artifact removal on ChIP quality metrics in ChIP-seq and ChIP-exo data. Front. Genet.5, 75 (2014).
- Li Q , BrownJB , HuangH , BickelPJ. Measuring reproducibility of high-throughput experiments. aoas5, 1752–1779 (2011).
- Yu G , WangL-G , HeQ-Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics31, 2382–2383 (2015).
- Nordin A , ZambaniniG , PagellaP , CantùC. The CUT&RUN blacklist of problematic regions of the genome. bioRxiv doi: 10.1101/2022.11.11.516118 (2022).
- Patro R , DuggalG , LoveMI , IrizarryRA , KingsfordC. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods14, 417–419 (2017).
- Love MI , HuberW , AndersS. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 550 (2014).
- Wang S , SunH , MaJet al. Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat. Protoc.8, 2502–2515 (2013).
- Wu T , HuE , XuSet al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb.)2, 100141 (2021).
- Xie Z , BaileyA , Kuleshovet al. Gene set knowledge discovery with Enrichr. Curr. Protoc1, e90 (2021).
- Supek F , BošnjakM , ŠkuncaN , ŠmucT. REVIGO summarizes and visualizes long lists of gene ontology terms. PLOS ONE6, e21800 (2011).
- Krämer A , GreenJ , PollardJJr , TugendreichS. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics30, 523–530 (2014).
- Gamble MJ , FrizzellKM , YangC , KrishnakumarR , KrausWL. The histone variant macroH2A1 marks repressed autosomal chromatin, but protects a subset of its target genes from silencing. Genes Dev.24, 21–32 (2010).
- Gaspar-Maia A , QadeerZA , HassonDet al. MacroH2A histone variants act as a barrier upon reprogramming towards pluripotency. Nat. Commun.4, 1565 (2013).
- Changolkar LN , SinghG , CuiKet al. Genome-wide distribution of macroH2A1 histone variants in mouse liver chromatin. Mol. Cell. Biol.30, 5473–5483 (2010).
- Hurtado-Bagès S , PosavecMarjanovic M , ValeroVet al. The histone variant MacroH2A1 regulates key genes for myogenic cell fusion in a splice-isoform dependent manner. Cells9, doi: 10.3390/cells9051109 (2020).
- Recoules L , HeurteauA , RaynalFet al. The histone variant macroH2A1.1 regulates RNA polymerase II-paused genes within defined chromatin interaction landscapes. J. Cell Sci.135, doi: 10.1242/jcs.259456 (2022).
- Bernstein E , Muratore-SchroederTL , DiazRLet al. A phosphorylated subpopulation of the histone variant macroH2A1 is excluded from the inactive X chromosome and enriched during mitosis. Proc. Natl Acad. Sci. USA105, 1533–1538 (2008).
- Mermoud JE , CostanziC , PehrsonJR , BrockdorffN. Histone macroH2A1.2 relocates to the inactive X chromosome after initiation and propagation of X-inactivation. J. Cell Biol.147, 1399–1408 (1999).
- Chadwick BP , WillardHF. Histone H2A variants and the inactive X chromosome: identification of a second macroH2A variant. Hum. Mol. Genet.10, 1101–1113 (2001).
- Kustatscher G , HothornM , PugieuxC , ScheffzekK , LadurnerAG. Splicing regulates NAD metabolite binding to histone macroH2A. Nat. Struct. Mol. Biol.12, 624–625 (2005).
- Son MJ , SonM-Y , SeolBet al. Nicotinamide overcomes pluripotency deficits and reprogramming barriers. Stem Cells31, 1121–1135 (2013).
- Meng Y , RenZ , XuFet al. Nicotinamide promotes cell survival and differentiation as kinase inhibitor in human pluripotent stem cells. Stem Cell Reports11, 1347–1356 (2018).
- Teslaa T , TeitellMA. Pluripotent stem cell energy metabolism: an update. EMBO J.34, 138–153 (2015).
- Posavec Marjanović M , Hurtado-BagèsS , LassiMet al. MacroH2A1.1 regulates mitochondrial respiration by limiting nuclear NAD consumption. Nat. Struct. Mol. Biol.24, 902–910 (2017).
- Hu Y , ChopraV , ChopraRet al. Transcriptional modulator H2A histone family, member Y (H2AFY) marks Huntington disease activity in man and mouse. Proc. Natl Acad. Sci. USA108, 17141–17146 (2011).
- Kapadia M , MianMF , MaDet al. Differential effects of chronic immunosuppression on behavioral, epigenetic, and Alzheimer’s disease-associated markers in 3xTg-AD mice. Alzheimers Res. Ther.13, 30 (2021).
- Haremaki T , MetzgerJJ , RitoT , OzairMZ , EtocF , BrivanlouAH. Self-organizing neuruloids model developmental aspects of Huntington’s disease in the ectodermal compartment. Nat. Biotechnol.37, 1198–1208 (2019).
- Ryskamp DA , KorbanS , ZhemkovV , KraskovskayaN , BezprozvannyI. Neuronal sigma-1 receptors: signaling functions and protective roles in neurodegenerative diseases. Front. Neurosci.13, 474181 (2019).
- Vadodaria KC , JonesJR , LinkerS , GageFH. Modeling brain disorders using induced pluripotent stem cells. Cold Spring Harb. Perspect. Biol.12, doi: 10.1101/cshperspect.a035659 (2020).
- Curran T , MorganJI. Fos: an immediate-early transcription factor in neurons. J. Neurobiol.26, 403–412 (1995).
- Krendl C , ShaposhnikovD , RishkoVet al. GATA2/3-TFAP2A/C transcription factor network couples human pluripotent stem cell differentiation to trophectoderm with repression of pluripotency. Proc. Natl Acad. Sci. USA114, E9579–E9588 (2017).
- Dowen JM , FanZP , HniszDet al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell159, 374–387 (2014).
- Levine M . Paused RNA polymerase II as a developmental checkpoint. Cell145, 502–511 (2011).
- Shu J , ZhangK , ZhangMet al. GATA family members as inducers for cellular reprogramming to pluripotency. Cell Res.25, 169–180 (2015).
- Song Y , LiangZ , ZhangJet al. CTCF functions as an insulator for somatic genes and a chromatin remodeler for pluripotency genes during reprogramming. Cell Rep.39, 110626 (2022).
- Schuster J , de GuidiC , FatimaA , SobolM , DahlN. Syndromic RNA polymerase II insufficiency: Generation of a human induced pluripotent stem cell line (UUIGPi002A-5) with a heterozygous disruption of POLR2A. Stem Cell Res.57, 102577 (2021).
- Markov GJ , MaiT , NairSet al. AP-1 is a temporally regulated dual gatekeeper of reprogramming to pluripotency. Proc. Natl Acad. Sci. USA118, doi: 10.1073/pnas.2104841118 (2021).
- Hu D , AbbasovaL , SchilderBM , NottA , SkeneNG , MarziSJ. CUT&Tag recovers up to half of ENCODE ChIP-seq peaks in modifications of H3K27. bioRxiv doi: 10.1101/2022.03.30.486382 (2023).
- Volpato V , SmithJ , SandorCet al. Reproducibility of molecular phenotypes after long-term differentiation to human iPSC-derived neurons: a multi-site omics study. Stem Cell Reports11, 897–911 (2018).
- Volpato V , WebberC. Addressing variability in iPSC-derived models of human disease: guidelines to promote reproducibility. Dis. Model. Mech.13, doi: 10.1242/dmm.042317 (2020).
- Carcamo-Orive I , HoffmanGE , CundiffPet al. Analysis of transcriptional variability in a large human iPSC library reveals genetic and non-genetic determinants of heterogeneity. Cell Stem Cell20, 518–532.e9 (2017).
- Silva M , DaheronL , HurleyHet al. Generating iPSCs: translating cell reprogramming science into scalable and robust biomanufacturing strategies. Cell Stem Cell16, 13–17 (2015).