Abstract
Three new phloroglucinol type compounds Indigoferin-A (1), Indigoferin-B (2) and Indigoferin-C (3), along with a known compound β-sitosterol were isolated from the Indegofera gerardiana Wall. The structures of Indigoferin-A (1), Indigoferin-B (2), and Indigoferin-C (3) were deduced on the basis of spectroscopic techniques (EI-MS, HREI-MS, 1H NMR, 13C NMR, HMQC, and HMBC). The urease inhibition studies on all the four compounds have also been carried out.
Introduction
The plant Indigofera gerardiana Wall., belongs to the family Leguminoseae (Fabeaceae)Citation1. While distributed worldwide, in Pakistan, it is found in mountainous areas of Khyber Pukhtoonkhwa, Azad Kashmir, and northern areas, from 1500 to 3000 metersCitation1. In India, it is found in the Himalayas. It is also found in Kasi Afghanistan and Western ChinaCitation2. I. gerardiana finds various medicinal uses in the indigenous systems of medicine. In northern areas of Pakistan, this plant is traditionally used for hepatitis, whooping cough, as antispasmodic, and as tonicCitation1,Citation3,Citation4. The extract of the plant prevents the development of hypoglycemia in mouse. The leaves, flowers, and tender shoots have the cooling, demulcent action and are also used in the treatment of leprosy and cancerous infectionsCitation5. The leaves are applied to abscesses whereas the roots are chewed to heal toothache and apathyCitation4. The alcoholic extract of the dried shoots exhibit the anti-inflammatory activityCitation6. The root bark, when chewed, gives relief in case of colic and skin infections involving microorganismsCitation7. The leaves, barks, and roots also showed antibacterial activityCitation4,Citation8.
Urease (urea amidohydrolase, EC: 3.5.1.5) occurs throughout the animal and plant kingdom. Many microorganisms use this enzyme as a source of nitrogen for growth, and it also plays an important role in plant nitrogen metabolism during the germination processCitation9,Citation10. The presence of urease activity in soils is exploited in the widespread agriculture. Unfortunately, excessive levels of soil urease can degrade fertilizer’s urea too rapidly and result in phytopathic effects and loss of volatilized ammoniaCitation11. However, in medical and veterinary interest, urease is a virulence factor in certain human and animal pathogens; it participates in the development of kidney stones, pyelonephritis, peptic ulcers, and other disease statesCitation12. The obvious remedy for treating bacterial infection with antimicrobials has often proven futileCitation13, and only a few combination regiments have reached clinical practice. Thus the need for alternative or novel treatment is evident. The discovery of potent and safe urease inhibitors have been a very important area of pharmaceutical research due to the involvement of ureases in different pathological conditions.
In the present study, we report herein the isolation and structure determination of three new compounds Indigoferin-A (1), Indigoferin-B (2), and Indigoferin-C (3), along with a known compound, β-sitosterol, and their urease inhibition activity.
Methods and materials
General experimental procedures
Optical rotations were measured on a JASCO DIP 360 polarimeter. IR spectra were recorded on a JASCO 320-A spectrophotometer. UV spectra were recorded on a HITACHI U-3200 spectrophotometer. EI-MS and high-resolution electron impact mass spectrum (HREI-MS) were recorded on Finnegan MAT 311 with MASPEC Data system and on JMS-HX 110 mass spectrometers. The 1H and 13C NMR spectra were recorded on Bruker, AM-300, AM-400, and AMX-500 NMR spectrometers operating at 400 MHz, (100 MHz for 13C). The chemical shifts values are reported in ppm (δ) units, and the coupling constants (J) are given in Hz.
Chromatographic conditions
For thin layer chromatography (TLC), precoated aluminum sheets (silica gel 60F-254, E. Merck) were used. Visualization of the TLC plates was achieved under UV at 254 and 366 nm and by spraying with cerric sulphate reagent. Solvent systems, “ethyl acetate-methanol; 9.3:0.7, 9.7:0.3,” were used.
Plant material
I. gerardiana Wall. were collected from upper Dir, Khyber Pukhtoonkhwa (Pakistan), during the month of April 2005. The plant was identified by Prof. Dr. Jahandar Shah, plant taxonomist, University of Malakand, Chakdara. The voucher specimen number GI-014 was placed in the herbarium of botany department, University of Malakand, Chakdara, Dir (L), Pakistan.
Extraction and isolation
The plant sample with an appropriate quantity was obtained after the preliminary necessary preparations such as drying under shadow for 3 weeks then comminuted into fine particles and pulverized into fine powder. The grinded plant (10 kg) was soaked in MeOH (80% v/v) with rare stirring at room temperature. After 2 weeks, the materials dissolved in MeOH were separated through the process of filtration. The procedure was replicated three times, and the clear material obtained was converted into syrupy liquid in vaco at 40°C to bestow dark brown paste. The crude MeOH extract (463.5 g) was suspended in distilled water and extracted with n-hexane (20.71% w/w), chloroform (15.96% w/w), ethyl acetate (12.94%), and n-butanol (19.41% w/w), and finally the aqueous (30.96% w/w) fraction was obtained. Each organic extract was then evaporated to dryness.
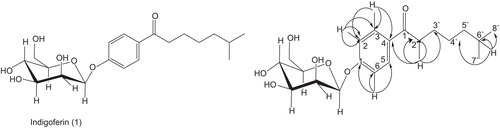
Indigoferin-A (1)
Black gummy solid; Rf 0.28 [methanol: chloroform; 3:97]; UV(EtOH) λmax (log ϵ) 575 nm; IR υmaxCHCl3, 35.21(OH groups), 1687 (keto-carbonyl), 1H-NMR (400 MHz, MeOD): 13C-NMR (MeOD, 100MHz) (see ); EIMS (M+ m/z): 382.4487 (observed), C20H30O7, 382.4481 (calculated).
Table 1. 1H (400 MHz) and 13C NMR (100 MHz) data of compound 1–3 in MeOD.
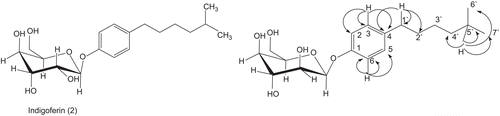
Indigoferin-B (2)
Yellow amorphous powder; yield 1.94 × 10−3%; Melting point 140–142°C; Rf 0.36 [methanol: chloroform; 7:93]: IRνmax cm−1 (CH3OH), 6545-3402 (OH groups), 1690 (keto-carbonyl); UV λmax (MeOH), 556 nm; 1H-NMR (400 MHz, MeOD); 13C-NMR (CDCl3, 100 MHz); HREI-MS (m/z), 268.3066 (observed), C14H20O5, 268.3061 (calculated).
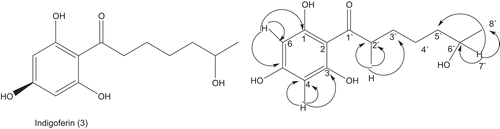
Indigoferin-C (3)
Brown powder, yield, 3.0 × 10−3%; melting point 145–147°C; Rf 0.40 [methanol: chloroform; 7:93]; IRνmax cm−1 (CH3OH), 2922 (C-H), 1623-1517 (aromatic C=C), 1230 (C=O); UV λmax (MeOH), 577 nm; 1H-NMR (400 MHz, MeOD), 13C-NMR (MeOD, 100MHz); HREI-MS (m/z), 354.4384 (observed), C19H30O6, 354.4380 (calculated).
Urease assay and inhibition
Reaction mixtures comprising 25 μL of enzyme (jack bean urease) solution and 55 μL of buffers containing 100 mM urea were incubated with 5 μL of test compounds (0.5 mM concentration) at 30°C for 15 min in 96-well plates. Urease activity was determined by measuring ammonia production using the indophenol method as described by weather burnCitation14. Briefly, 45 μL each phenol reagent (1% w/v phenol and 0.005% w/v sodium nitroprusside) and 70 μL of alkali reagent (0.5% w/v NaOH and 0.1% active chloride NaOCl) were added to each well. The increasing absorbance at 630 nm was measured after 50 min, using a microplate reader (Molecular Device). All reactions were performed in triplicate in a final volume of 200 μL. The results (change in absorbance per minute) were processed by using softMax Pro software (molecular Device). The entire assays were performed at pH 6.8. Percent inhibitions were calculated from the formula 100-(ODtestwell/ODcontrol) × 100. Thiourea was used as the standard inhibitor of ureaseCitation15,Citation16.
Results and discussion
Indigoferin-A (1)
Compound 1 was obtained from the ethyl acetate fraction of the methanolic extracts of I. gerardiana as black gummy solid. The EI-MS spectrum of compound 1 showed the molecular ion peak [M+] at m/z 382. The molecular formula C20H30O7 was establish by HREI-MS, giving the molecular ion peak at m/z 382.447 (calculated for C20H30O7, 382.4481), having six degrees of unsaturation. The IR spectrum of compound 1 showed absorption band at 3521 and 1687 cm−1 for hydroxyl and keto carbonyl groups.
The 1H NMR spectrum of compound 1 showed signal for methyl, methylene, and methine protons. In the upfiled region of the spectrum, two doublets each of three protons integration at δ 0.87 and 0.89 with coupling constants 6.6 Hz (each) were assigned to the terminal methyl protons. Similarly, a multiplet of one proton at δ 1.46 was assigned to C-6′ methine proton. Two double doublets, each of one proton integration at δ 3.00 and 2.97 having coupling constants 15.4, 6.6 Hz and 15.4, 7.1 Hz, were assigned to the C-2′ methylene protons. In the downfield region, signals for aromatic protons at δ 7.03–7.83 were also observed, while a doublet of one proton at δ 5.50 with a coupling constant 7.4 Hz was assigned to the C-1″ anomeric proton. Similarly, two broad doublets, each of one proton integration at δ 4.40 and 4.52 with coupling constant J 11.7 Hz (each) were assigned to C-6″ methylene protons of sugar moiety. The glucose was recognized as β-d-glucose by means of its 1H- and 13C-NMR data (). The acid hydrolysis of compound 1 provided glycone, which was separated and identified as D-glucose.
The 13C NMR spectrum (BB, DEPT; ) of compound 1 showed 20 signal, including 2 methyl, 5 methylene, 10 methine, and 3 quaternary carbons. The 1H- 13C-correlation was determined by HMQC spectrum. Similarly the extended range 1H- 13C connectivities were obtained through HMBC technique (). In the HMBC spectrum H-6′ proton (δ 1.46), showed correlations with C-5′ (δ 39.5), C-7′ (δ 21.9), C-8′ (δ 21.0) and C-6′ (δ 38.0), while H-2′ (δ 3.00) showed its correlations with C-1′ (δ 203.5), C-4 (δ 131.6), C-2′ δ 39.0), and C-3′ (δ 25.2). Likewise, H-1″ proton (δ 5.50) showed its correlation with C-2″ (δ 72.5) and C-1 (δ 158.4).
On the basis of above spectral evidences, the structure of compound 1 was established as Indigoferin-A [(6-methyl-1-(4-((2S,3S,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yloxy)phenyl)heptan-1-one)].
Indigoferin-B (2)
Compound 2 was obtained from the ethyl acetate fraction of the methanolic extracts of I. gerardiana as brown powder. The EI-MS spectrum of compound 2 showed molecular ion peak [M+] at m/z 354. The molecular formula C19H30O6 was customized by HREI-MS, exhibiting the molecular ion peak at m/z 354.4384 (calculated for C19H30O6, 354.4380), having five degree of unsaturation. The IR spectrum of compound 2 showed absorption bands at 2922 (C-H), 1623, 1517 (aromatic C=C), and 1230 (C-O) cm−1.
The 1H NMR spectrum of compound 2 showed very close resemblance to that of compound 1. In the upfield region of the spectrum, two doublets each of three protons integration at δ 0.87 and 0.91 having coupling constant 6.8 Hz (each) were assigned to methyl protons. The signals for sugar moiety was observed at δ 5.46 (1H, d, J = 7.2 Hz) which may be assigned to anomeric proton H-1″, while four multiplets, each of single H integration at δ 4.21, 4.24, 3.87, and 3.99, were assigned to the methine protons of sugar moiety. The only difference in the spectrum of compound 2 and compound 1 is the absence of the signals at δ 3.00 and 2.97 in the spectrum of compound 2, indicating the absence of the carbonyal group in compound 2.
The 13C NMR spectrum (BB, DEPT; ) of compound 2 showed 19 signals, including 2 methyl, 4 methylene, 10 methine, and 2 quaternary carbons. The 1H- 13C- correlations were determined by HMQC spectrum, whereas the long range 1H- 13C-connectivities were established through HMBC technique.
In the HMBC spectrum (), H-5′ (δ 1.46), showed correlation with C-4′, C-6′, and C-7′, while H-1′ (δ 2.61) showed its correlation with C-4 (δ 135.5), C-1′, (δ 35.5), and C-2′ (δ 31.1). Similar H-1″ (δ 5.46) showed its correlation with C-2″ (δ 74.9) and C-1 (δ 157.2).
Based on the above spectral data, the structure of compound 2 was established as Indigoferine-B, (2R,3R,4R,5R,6S)-2-(hydroxymethyl)-6-(4-(5-methylhexyl)phenoxy)tetrahydro-2H-pyran-3,4,5-triol.
Indigoferin-C (3)
Compound 3 was secluded from the EtOAc soluble part of the of I. gerardiana as yellow amorphous powder. In EI-MS spectrum of compound 3, the molecular ion peak was observed at m/z 268. The high-resolution electron impact mass spectrum (HREI-MS) customized the molecular formula C14H20O5, viewing M+ peak at m/z 268.3066 (calculated for C14H20O5, 268.3061), having five degree of unsaturation. The IR spectrum of compound 3 showed the bands at 3545 and 1690 cm−1 for hydroxyl and carbonyl functionalities, respectively.
The 1H NMR spectrum of compound 3 illustrated signals for methyl, methylene, and methine protons. In the downfield region of the spectrum, two doublets each of one proton integration at δ 6.15 and 5.80 having the coupling constant 2.2 Hz were assigned to the aromatic protons. The upfield shifts and the smaller coupling constant of these protons indicated that these meta coupled protons are between oxygenated quartenary carbons16. In the upfield region of the spectrum, a doublet of the three protons at δ 1.09 with coupling constant 6.6 Hz was assigned to the methyl protons.
The 13C NMR spectrum (Broad-Bond BB, DEPT; ) of compound 3 indicated 14 signals, including 1 methyl, 5 methylenes, 3 methine, and 5 quaternary carbons. The 1H- 13C relationships are determined by HMQC spectrum, whereas the long-range 1H- 13C-connectivities were obtained through HMBC technique ().
In the HMBC spectrum (), H-2′ (δ 3.09), showed 1J and 2J relationship with C-1′ (δ 208.1), C-2′ (δ 42.8), C-3′ (δ 24.9), and C-2 (δ 106.5). Similarly, H-6′ (δ 1.89) showed its correlation with C-5′ (δ 34.5), C-6′ (δ 38.0), C-7′ (δ 68.5), and C-8′ (δ 17.2). The H-4 (δ 5.80), exhibited relationship with C-3 (δ 164.4), C-4 (δ 95.4), C-5 (δ 165.6) and C-2 (δ 106.5). On the basis of the above spectral evidences, the structure of the compound 3 was establish as Indigoferin-C.
Organic compound isolated from higher plants have extensive past and present use in the treatment of many diseases. So these naturally isolated compounds were biologically screened for their inhibitory activities against urease. Two compound, namely, compounds 2 and 3 exhibited excellent in vitro inhibitory activities having IC50 value 23.33 ± 0.11 and 49.7 ± 0.40 μM, respectively, as compared with standard thiourea which have IC50 value 21 ± 0.011 μM. Whereas, compound 1 showed no activity (). The comparison inhibitory activities of compounds 1, 2, and 3 indicated that the aliphatic chain is responsible for activities of the compounds. These compounds showed different activities possibly due to the difference in their aliphatic chain. Compound 2 and 3 possess aliphatic chain of shorter length as compared with compound 1. When the aliphatic chain increased by one carbon as in compound 1, the compound become inactive. The results are summarized in .
Table 2. Tabular representation of urease inhibitory activity of compounds 1–4.
Acknowledgements
We are grateful to Prof. Dr. Jahan Dar Shah, Department of Botany, University of Malakand, Chkadara, Dir (L), Khyber Pukhtoonkhwa, Pakistan, for helping in identification of the plant.
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Shinwari ZK, Rehman M-U Watanabe, T, Yoshikawa T. A pictorial guide to medicinal plants of Pakistan. 2006; Kohat, Pakistan: Kohat University of Science and Technology.
- Anonymous. The wealth of India. A Dictionary of Indian Raw Materials & Industrial Products. Volume-V, Council of Scientific & Industrial Research, New Delhi, India Publications, New Delhi, India, pp 179–180 & 230 (2001).
- Khan MH, Khan MA. Common medicinal folk recipes of District Buner, NWFP, Pakistan. J Ethnobot Leaflets.2003.
- Gamble JS. A Manual of Indian Timbers. Bishen Singh Mahendra Pal Singh (1972).
- Nyarko AK, Sittie, AA. Safety evaluation of an antidiabetic plant extract in non-diabetic human volunteers. J Phytotherapia Research Indigofera Arrecta 1998;12:52–54.
- Amala Bhaskar E, Ganga N, Arivudainambi R, Santhanam G. Anti-inflammatory activity of Indigofera aspalathoides Vahl. Indian J Med Res 1982;76 Suppl:115–118.
- Esimon COA, Dikwu MU, Muko KN. Antibacterial properties of Indigofera dendroides leaves. Fitroterapia 1999;70:517–520.
- Umar M. Antibacterial and antifungal activity of small protein of Indigofera oblongifolia leaves. J Ethanopharmacol 1999;64:277–282.
- Mobleyand HLT, Hausinger, RP. Microbial urease: Significance, regulation and molecular characterization. Rev 1989;53:85–108.
- Zonia LE, Stebbins NE, Polacco JC. Essential role of urease in germination of nitrogen-limited Arabidopsis thaliana seeds. Plant Physiol 1995;107:1097–1103.
- Mulvaney RL, Bremner JM. in Soil Biochemistry (E. A., Paul, and J.N. Ladd, Eds.) (1981) pp 153–196, Marcel Dekker, Inc., New York.
- Mobley HLT, Island MD, Hausinger RP. Molecular biology of microbial ureases. Microbiol Rev 1995;59:451–480.
- Bayerdörffer E, Ottenjann R. The role of antibiotics in Campylobacter pylori associated peptic ulcer disease. Scand J Gastroenterol Suppl 1988;142:93–100.
- Kosasi S, Sluis WGVG, Labudie RP. Phytocemistry 1989;28:2439.
- Khan KM, Iqbal S, Lodhi MA, Maharvi GM, Ullah Z, Choudhary MI et al. Biscoumarin: new class of urease inhibitors; economical synthesis and activity. Bioorg Med Chem 2004;12:1963–1968.
- Weatherburn MW. Anal Chem 1967;39:971.