Abstract
Polypyridyl chlororuthenium(II) complexes have been synthesized and characterized. The binding mode of the complexes to DNA has been evaluated from the combined results of electronic absorption spectroscopy and viscosity measurement study. The results suggest that complexes 1, 2 and 3 bind to DNA via classical intercalation, electrostatic interaction and partial intercalation mode, respectively. Complex 2 shows less affinity for DNA. Cleavage of pUC19 DNA by complexes has been checked using gel electrophoresis. The data disclose that complex 1 has the highest cleaving ability.
Introduction
An interaction of polypyridyl ruthenium complexes with DNA has attracted a considerable attention in recent decades for developing novel probes of DNA structure or new therapeutic agents.Citation1–3 Ruthenium(II) complexes bind to DNA in a non-covalent interaction fashion, such as electrostatic binding for cation, groove binding for large ligands, intercalative binding for planar ligands and partial intercalative binding for incomplete planar ligands.Citation4–6 Several ruthenium complexes were found to be potent anticancer substances with remarkable activity and lower toxicity than platinum complexes.Citation7
In co-ordination chemistry, terpyridines are of special interest due to their ability to form stable complexes with many transition metal ions. Such complexes possess interesting photophysical, electrochemical and photochemical properties, allowing constructing extended supramolecular architectures.Citation8 The influence of the ancillary ligands of the complexes has received little attention. Since the octahedral polypyridyl RuII complexes bind to DNA in three dimensions, the ancillary ligands can also play an important role in governing DNA binding of complexes. At the same time, varying substitutive group or substituent position in the ancillary ligand can also create some interesting differences in the space configuration and the electron density distribution of RuII polypyridyl complexes, which will result in some differences in spectral properties, the DNA-binding behaviours of the complexes and will also be helpful to understand the binding mechanism of RuII polypyridyl complexes to DNA.Citation9–13
In a previous publication from our laboratory,Citation14 biological activities of copper complexes with gatifloxacin and various neutral bidentate ligands were studied. In this article, we reported the synthesis and characterization of [RuII(4-bptpy)(dmphen)Cl]ClO4, [RuII(4-fptpy)(dmphen)Cl]ClO4 and [RuII(4-mptpy)(dmphen)Cl]ClO4 complexes. Binding affinity of the complexes towards Herring Sperm DNA has been quantified by calculating binding constant (Kb), using absorption titration methodology. The binding mode of the complexes with DNA has been determined using viscosity measurements. Cleavage ability of complexes towards pUC19 DNA has also been investigated by gel electrophoresis technique.
Experimental
Materials and instrumental details
2-Acetyl pyridine, 4-bromobenzaldehyde, 4-fluorobenzaldehyde and 4-methoxybenzaldehyde were purchased from Spectrochem (Mumbai, India). Ruthenium trichloride and sodium perchlorate were purchased from Chemport (Mumbai, India). Agarose, ethidium bromide (EB), TAE (Tris–acetyl–EDTA), bromophenol blue and xylene cyanol FF were purchased from Himedia (Mumbai, India). Herring Sperm DNA was purchased from Sigma Chemical Co. (Nashik, India). 2,9-Dimethyl-1,10-phenanthroline was purchased from Loba Chemie (Mumbai, India). Infrared spectra were recorded on Fourier transform IR (FTIR) Shimadzu spectrophotometer as KBr pellets in the range 4000–400 cm−1. The 1H and 13C NMR were recorded on a Bruker Avance (400 MHz). The fast atomic bombardment mass spectra (FABMS) were recorded on Jeol SX 102/Da-600 mass spectrophotometer/data system using argon/xenon (6 kV, 10 mA) as the FAB gas. The accelerating voltage was 10 kV and spectra were recorded at room temperature. The electronic spectra were recorded on a UV-160A UV–vis spectrophotometer (Shimadzu, Japan). Thermogravimetric analysis (TGA) was carried out using a 5000/2960 SDTA (TA Instrument, New Castle, DE) operating at a heating rate of 10°C/min in the range of 20–800°C in N2. C, H and N elemental analyses were performed with a model 240 Perkin Elmer elemental analyzer.
Experimental procedure for the synthesis of terpyridines
4′-(4-Bromophenyl)-2,2′:6′,2′′-terpyridine (4-bptpy)
2-Acetylpyridine (2.42 g, 20.0 mmol) was added to an ethanolic solution of 4-bromobenzaldehyde (1.85 g, 10.0 mmol) in EtOH (70 mL). KOH pellets (1.4 g, 26 mmol) and aqueous NH3 (30 mL, 25%, 0.425 mol) were added to the solution, and the mixture was then stirred at room temperature for 8 h. An off-white solid formed was collected by filtration and washed with H2O (3 × 10 mL) and EtOH (2 × 5 mL). Crystallization from CHCl3–MeOH gave white crystalline solid. Yield: 1.84 g, 47.54%, mp: 125°C. 1H NMR (CDCl3, 400 MHz) δ/ppm 8.591–8.721 (m, 6H), 7.984 (t, 2H, H4,4′′), 7.802 (d, 2H, Hph2,6), 7.710 (d, 2H, Hph3,5), 7.486 (d, 2H, H5,5′′). 13C NMR (CDCl3, 100 MHz) δ/ppm 156.15 (C2′,6′), 155.27 (C2,2′′), 149.67 (C6,6′′), 148.53 (C4′), 137.77 (Cph1), 137.08 (C4,4′′), 132.66 (C3,3′′), 129.35 (Cph2,6), 124.89 (C5,5′′), 123.48 (Cph4), 121.34 (Cph3,5), 118.14 (C3′,5′). Anal. Calc. for C21H14N3Br: C 64.96, H 3.63, N 10.82. Found: C 64.76, H 3.83, N 10.67%.
4′-(4-Fluorophenyl)-2,2′:6′,2′′-terpyridine (4-fptpy)
This ligand was prepared by the same method described above, but using 4-fluorobenzaldehyde instead of 4-bromobenzaldehyde. Yield: 1.32 g, 40.49%, mp: 182°C. 1H NMR (CDCl3, 400 MHz) δ/ppm 8.691–8.743 (m, 6H), 7.902–7.918 (m, 4H, Hph2,3,5,6), 7.391 (t, 2H, H4,4′′), 7.215 (t, 2H, H5,5′′). 13C NMR (CDCl3, 100 MHz) δ/ppm 162.29 (Cph4), 155.96 (C2′,6′), 155.73 (C2,2′′), 149.40 (C4′), 148.94 (C6,6′′), 137.18 (C4,4′′), 129.22 (C3,3′′), 129.12 (Cph2,6), 123.97 (C5,5′′), 121.52 (Cph1), 118.91 (C3′,5′), 115.82 (Cph3,5). Anal. Calc. for C21H14N3F: C 77.05, H 4.31, N 12.84. Found: C 77.24, H 4.09, N 12.71%.
4′-(4-Methoxyphenyl)-2,2′:6′,2′′-terpyridine (4-mptpy)
This ligand was prepared by the same method described above but using 4-methoxybenzaldehyde instead of 4-bromobenzaldehyde. Yield: 1.16 g, 34.31%, mp: 158°C. 1H NMR (CDCl3, 400 MHz) δ/ppm 8.76–8.795 (m, 4H, H3,3′,5′,3′′), 8.71 (d, 2H, H6,6′′), 7.913–7.940 (m, 4H), 7.396 (d, 2H, Hph3,5), 7.055 (dd, 2H, H5,5′′), 3.90 (s, 3H, OCH3). 13C NMR (CDCl3, 100 MHz) δ/ppm 160.57 (Cph4), 156.15 (C2′,6′), 155.59 (C2,2′′), 149.81 (C4′), 148.88 (C6,6′′), 137.08 (C4,4′′), 130.63 (Cph1), 128.55 (Cph2,6), 123.81 (C5,5′′), 121.48 (C3,3′′), 118.41 (C3′,5′), 114.35 (Cph3,5), 55.38 (OCH3). Anal. Calc. for C22H17N3O: C 77.86, H 5.05, N 12.38. Found: C 77.65, H 4.78, N 12.53%.
Experimental procedure for the synthesis of RuII complexes
[RuII(4-bptpy)(dmphen)Cl]ClO4 (1)
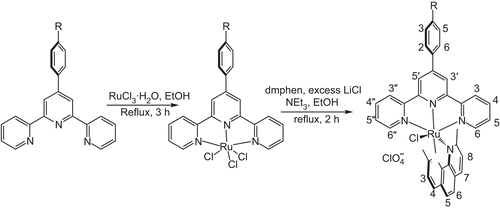
[RuIII(4-bptpy)Cl3] was synthesized by a method described previously.Citation15 [RuII(4-bptpy)(dmphen)Cl]ClO4 (1) was synthesized by taking [RuIII(4-bptpy)Cl3] (262 mg, 0.44 mmol), 2,9-dimethyl-1,10-phenanthroline (104 mg, 0.5 mmol), excess LiCl (122 mg, 2.94 mmol) and NEt3 (0.9 mL) in 45 mL of ethanol, and the mixture was refluxed for 2 h under a dinitrogen atmosphere (). The initial dark brown colour of the solution gradually changed to a deep purple. The solvent was then removed under reduced pressure. The dry mass was dissolved in a minimum volume of acetonitrile, and an excess saturated aqueous solution of NaClO4 was added to it. The precipitate was filtered off and washed with cold ethanol followed by ice-cold water. The product was dried in vacuum and purified using a silica column. The complex was eluted by 2:1 CH2Cl2/CH3CN. Yield: 0.231 g, 63%, mol. wt. 832.39. IR (KBr): ν 3063 w,br; 2922 sh; 1596 m,sh; 1498 m,sh; 1086 s,sh; 754 s,sh; 627 vs,sh; 516 w,sh cm−1. 1H NMR [dimethyl sulphoxide-d6 (DMSO-d6), 400 MHz] δ/ppm 9.512 (s, 2H, T3′,5′, where T = terpyridine), 9.116 (d, 2H, T6,6′′), 8.441 (d, 2H, P4, where P = phenanthroline), 8.428 (d, 2H, T3,3′′), 8.111 (d, 2H, Tph3,5), 8.084 (t, 2H, T4,4′′), 7.866 (s, 2H, P5,6), 7.698 (d, 1H, P7), 7.621 (d, 1H, P3), 7.583 (d, 1H, P8), 7.561 (d, 2H, Tph2,6), 7.292 (t, 2H, T5,5′′), 3.36 (s, 3H, CH3), 2.78 (s, 3H, CH3). Anal. Calc. for C35H26N5O4BrCl2Ru: C 50.50, H 3.15, N 8.41. Found: C 50.35, H 3.27, N 8.28%. FABMS: m/z = 209 [dmphen + H]+, 699 [M− ClO4− Cl]+, 734 [M− ClO4]+, 735 [M− ClO4 + H]+, 833 [M]+.
[RuII(4-fptpy)(dmphen)Cl]ClO4 (2)
This complex was synthesized in a manner identical to that described for [RuII(4-bptpy)(dmphen)Cl]ClO4, with [RuIII(4-fptpy)Cl3] (235 mg, 0.44 mmol) in place of [RuIII(4-bptpy)Cl3]. Yield: 0.265 g, 78%, mol. wt. 771.59. IR (KBr): ν 3058 w,br; 2923 sh; 1599 m,sh; 1496 m,sh; 1088 s,sh; 757 s,sh; 626 vs,sh; 483 w,sh cm−1. 1H NMR (DMSO-d6, 400 MHz) δ/ppm 9.489 (s, 2H, T3′,5′), 9.111 (d, 2H, T6,6′′), 8.524 (q, 2H, T3,3′′), 8.445 (d, 2H, P4), 8.086 (t, 2H, T4,4′′), 7.861 (s, 2H, P5,6), 7.672 (d, 1H, P7), 7.655 (t, 2H, Tph3,5), 7.623 (d, 1H, P3), 7.586 (d, 1H, P8), 7.563 (d, 2H, Tph2,6), 7.295 (t, 2H, T5,5′′), 3.363 (s, 3H, CH3), 2.786 (s, 3H, CH3). Anal. Calc. for C35H26N5O4FCl2Ru: C 54.48, H 3.40, N 9.08. Found: C 54.63, H 3.23, N 9.24%. FABMS: m/z = 209 [dmphen + H]+, 637 [M− ClO4− Cl]+, 672 [M− ClO4]+, 673 [M− ClO4 + H]+, 771 [M]+.
[RuII(4-mptpy)(dmphen)Cl]ClO4 (3)
This complex was synthesized in a manner identical to that described for [RuII(4-bptpy)(dmphen)Cl]ClO4, with [RuIII(4-mptpy)Cl3] (241 mg, 0.44 mmol) in place of [RuIII(4-bptpy)Cl3]. Yield: 0.241 g, 70%, mol. wt. 783.62. IR (KBr): ν 3070 w,br; 2929 sh; 1603 m,sh; 1495 m,sh; 1259 s; 1082 s,sh; 764 s,sh; 625 vs,sh; 464 w,sh cm−1. 1H NMR (DMSO-d6, 400 MHz) δ/ppm 9.44 (s, 2H, T3′,5′), 9.11 (d, 2H, T6,6′′), 8.435 (d, 2H, P4), 8.446 (d, 2H, T3,3′′), 8.067 (t, 2H, T4,4′′), 7.863 (s, 2H, P5,6), 7.693 (d, 1H, P7), 7.618 (d, 1H, P3), 7.572 (d, 1H, P8), 7.55 (d, 2H, Tph2,6), 7.326 (d, 2H, Tph3,5), 7.281 (t, 2H, T5,5′′), 3.968 (s, 3H, OCH3), 3.354 (s, 3H, CH3), 2.776 (s, 3H, CH3). Anal. Calc. for C36H29N5O5Cl2Ru: C 55.18, H 3.73, N 8.94. Found: C 55.37, H 3.61, N 8.76%. FABMS: m/z = 209 [dmphen + H]+, 649 [M− ClO4− Cl]+, 684 [M− ClO4]+, 685 [M− ClO4 + H]+, 783 [M]+.
Caution: Perchlorate salts of metal complexes with organic ligands are potentially explosive; hence, only small amounts of the material should be prepared and handled with great care.
Absorption titration
The absorption titrations of RuII complexes in the buffer were performed by using a fixed complex concentration to which increments of the nucleic acid stock solution was done. Concentration of complex solutions employed was 20 µM. Influence of DNA on MLCT band of RuII complexes were measured via UV–vis absorbance spectra.Citation16–19 After addition of equivalent amount of DNA to reference cell, incubation for 10 min at room temperature was provided, followed by absorbance measurement. DNA-mediated hypochromism (decrease in absorbance) or hyperchromism (increase in absorbance) for test compounds were calculated. The intrinsic binding constant Kb was determined by making it subject in the following equationCitation13:
where [DNA] is the concentration of DNA in base pairs, the apparent absorption coefficient ϵa, ϵf and ϵb correspond to Aobs/[Ru], the extinction coefficient for the free complex and the extinction coefficient for the free complex in the fully bound form, respectively. In plots [DNA]/(ϵa− ϵf) versus [DNA], Kb is given by the ratio of slope to the y-intercept.
Viscosity study
Viscosity measurement was carried out using a Cannon-Ubbelohde viscometer maintained at a constant temperature of 27.0 (±0.1)°C in a thermostatic jacket. DNA samples with an approximate average length of 200 base pairs were prepared by sonication in order to minimize complexities arising from DNA flexibility.Citation20 Flow time was measured with a digital stopwatch with an accuracy of 0.01 sec. Each sample was measured thrice with a precision of 0.1 sec and an average flow time was calculated. Data are represented graphically as (η/η0)1/3 versus concentration ratio ([complex]/[DNA]),Citation21 where η is the viscosity of DNA in the presence of complex and η0 is the viscosity of DNA alone. Viscosity values were calculated from the observed flow time of DNA-containing solutions (t > 100 sec), corrected for the flow time of buffer alone (t0), η = t− t0.
Quantitative determination of pUC19 DNA cleavage by agarose gel electrophoresis
Cleavage of pUC19 DNA by RuII complexes was measured by the conversion of supercoiled pUC19 DNA to open circular and linear. Gel electrophoresis of pUC19 DNA was carried out in TAE buffer (0.04 M Tris–acetate, pH 8, 0.001 M EDTA). Fifteen microlitres of reaction mixture contains complex and 100 µg/mL plasmid DNA. Reactions mixture was incubated at 37°C. All reactions were quenched by addition of 3 µL loading buffer (0.25% bromophenol blue, 40% sucrose, 0.25% xylene cyanol FF and 200 mM EDTA). The aliquots were loaded directly on to 1% agarose gel bed and electrophoresed at 50 V in 1× TAE buffer. Gel was stained with 0.5 µg/mL EB and was photographed on a UV illuminator. After electrophoresis, the proportion of DNA in each fraction was estimated quantitatively from the intensity of the bands using AlphaDigiDoc™ RT (Version V.4.0.0 PC-Image software).
Results and discussion
TGA and electronic absorption analysis
TGA data of the complexes show no weight loss between 80°C and 180°C. So, there is an absence of co-ordinated or lattice water molecule. The electronic spectra of the complexes consist of three well-defined bands in the range 250–500 nm, similar to that observed for [Ru(dpphen)(terpy)Cl]PF6 reported by Yoshikawa et al.Citation22 The lowest energy absorption band (MLCT band) for complexes 1, 2 and 3 appeared at 490.5, 488.5 and 492 nm, respectively (). Changing the substitution on terpyridine from 4-fptpy to 4-mptpy, red shift of the MLCT band is observed. The two higher energy absorption bands appeared in the range 282.5 to 285.5 and 308 to 310 nm, which can be assigned to the ligand-centred transitions dmphen(π)→dmphen(π*) and terpy(π)→terpy(π*), respectively.Citation22
Table 1. Electronic spectral data for the ruthenium(II) complexes.
Infrared spectroscopy
The band appeared at ~722 cm−1 is due to C–H out of plan bending. The presence of perchlorate as a counter ion is confirmed by the very strong, broad band at ~1085 cm−1 and the strong, sharp band around 625 cm−1.Citation23 In the spectrum of complex 3, band at 1259 cm−1 is due to the asymmetric stretching of aromatic ether. A weak, broad band around 3060 cm−1 is a characteristic of aromatic C–H stretching, whereas a sharp band at 2925 cm−1 is a characteristic of C–H stretching of methyl group. Sharp bands with medium intensity appear around 1600 and 1495 cm−1, which is a characteristic of aromatic ring stretching. An intense, sharp band at ~760 cm−1, which is a characteristic of ring deformations and C–H out-of-plane deformations, appears as expected from a structure including aromatic rings. Weak, sharp band in the range 464 to 516 cm−1 is a characteristic of Ru–N stretching mode. A Ru–Cl stretching mode is expected to be in the region <400 cm−1.Citation24
1H NMR
On co-ordination with ruthenium ion, chemical shift of the T3′,5′, T6,6′′ and T4,4′′ protons shows large downfield due to metal-to-ligand π-back donation.Citation25 Tph2,3,5,6 of complexes 2 and 3 show upfield shift. Tph2,6 of complex 1 shows small upfield shift, whereas Tph3,5 shows downfield shift. Small upfield shift of T3,3′′ protons observed in all complexes. No considerable change observed in chemical shift of T5,5′′ protons. One methyl group of dmphen appeared in downfield than other due to the ring-current anisotropic effect exerted by terpyridine, experienced through space.
Electronic absorption titration
DNA binding of polypyridyl RuII complexes with DNA can be quantified by monitoring changes in the electronic spectra at MLCT band. Complex that binds to DNA through intercalation usually results in hypochromism and bathochromism. The extent of the hypochromism commonly parallels the intercalative binding strength.Citation26,Citation27 Complexes interact with DNA through electrostatic interaction show lower hypochromicity with no bathochromic shift.Citation25,Citation28 The electronic spectral traces of the complexes in absence and presence of Herring Sperm DNA are given in .
Figure 1. Electronic absorption spectra of (A) [RuII(4-bptpy)(dmphen)Cl]ClO4, (B) [RuII(4-fptpy)(dmphen)Cl]ClO4 and (C) [RuII(4-mptpy)(dmphen)Cl]ClO4 with increasing amount of DNA in phosphate buffer (Na2HPO4/NaH2PO4, pH 7.2). [Complex] = 20 µM, [DNA] = 0–16.6 µM with incubation period of 15 min at 37°C. Plots of [DNA]/(ϵa− ϵf) versus [DNA] for the titration of DNA with RuII complexes.
![Figure 1. Electronic absorption spectra of (A) [RuII(4-bptpy)(dmphen)Cl]ClO4, (B) [RuII(4-fptpy)(dmphen)Cl]ClO4 and (C) [RuII(4-mptpy)(dmphen)Cl]ClO4 with increasing amount of DNA in phosphate buffer (Na2HPO4/NaH2PO4, pH 7.2). [Complex] = 20 µM, [DNA] = 0–16.6 µM with incubation period of 15 min at 37°C. Plots of [DNA]/(ϵa− ϵf) versus [DNA] for the titration of DNA with RuII complexes.](/cms/asset/3f89c834-d951-4afd-ace0-bef862654273/ienz_a_570007_f0002_b.gif)
As the DNA concentration is increased, hypochromism is observed in the MLCT band of each complex, as shown in . For complex 1, the MLCT absorption band shifts from 490.5 to 503.5 nm with 23.9% hypochromism. For complexes 2 and 3, the MLCT transition bands exhibit red shifts of 0 and 2 nm, and hypochromism of 5.6% and 13.3%, respectively. The bathochromic shift of complexes 1 and 3 suggests that they may bind to DNA via classical intercalative mode. Absence of red shift at MLCT band of complex 2 suggests that the complex may electrostatically interact with DNA. Highest hypochromicity at MLCT band was observed for complex 1, which shows that the complex 1 interacts with DNA more strongly than others. More confirmation regarding binding mode of the complexes will be obtained from viscosity measurement. The intrinsic binding constants Kb of complexes 1, 2 and 3 were found to be 6.32 × 105, 6.57 × 103 and 1.85 × 104 M−1, respectively (). The Kb value of complex 1 is comparable with the classical intercalator [Ru(dmp)2(HPIP)]2+.Citation11 Complexes with electron-withdrawing group on ancillary ligand possess higher DNA-binding affinity than the one having electron-donating group.Citation5 So, complex 1 has higher Kb value than complex 3. In fact, complex 2 also has electron-withdrawing group, but has low binding constant. Electrostatic binding mode may be responsible for its lower affinity towards DNA.
Table 2. Electronic absorption data upon addition of Herring Sperm DNA.
Viscosity measurement
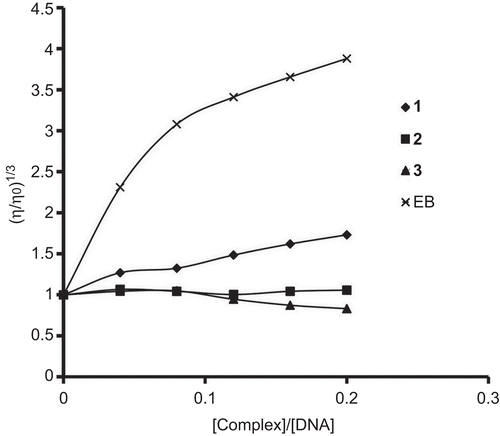
From viscosity measurement study, we confirmed the binding mode of complexes. A classical intercalation results in lengthening of the DNA helix, as base pairs are separated to accommodate the binding ligand, leading to increase in viscosity of DNA solution. A partial and/or non-classical intercalation of compound may bend DNA helix, resulting in the decrease of its effective length and thereby its viscosity.Citation13 When the compounds interact with DNA electrostatically, no effect on relative viscosity of DNA is observed.Citation25 The effect of increasing amount of EB and complexes on the relative viscosity of DNA is shown in . EB is a well-known classical intercalator. The relative viscosity of DNA solution increases with increase in amount of complex 1. So, complex 1 binds to DNA via classical intercalative mode. Complex [Ru(bpy)3]2+ has been known to bind DNA in electrostatic mode, and it exerts essentially no effect on DNA viscosity.Citation5 No change in relative viscosity was observed for complex 2, which led to a conclusion that complex 2 interacts with DNA electrostatically. Absorption titration also supports the same binding mode for complex 2 (0 nm red shift). In contrast, complex 3 decreases the relative viscosity of DNA as shown by the partial intercalators. Considering the results of spectroscopic and viscosity measurements, we suggested that complex 1 binds to DNA via classical intercalative mode, complex 2 interacts electrostatically and complex 3 partially intercalates to DNA.
Quantitative determination of pUC19 DNA cleavage by agarose gel electrophoresis
The double-stranded pUC19 DNA exists in a compact supercoiled (SC) conformation. If one strand breaks, the SC form of DNA will relax to produce an open circular (OC) form. If both strands are cleaved, a linear (L) form will be produced. Increasing order of the migration rate for all the three form of DNA is SC > L > OC.Citation29 DNA cleavage ability of the complexes was quantified by measuring the transformation of the SC form into OC and L forms. The concentration-dependent DNA cleavage induced by complex 1 with incubation time of 180 min is shown in . The percentage of different forms of DNA produced by various concentration of complex 1 is shown in . The data indicate that as the concentration of complex increases, percentage of OC and L forms increases. The linear form generates when concentration is ≥125 µM.
Figure 3. Agarose gel (1%) of pUC19 (100 µg/mL) incubated for 2 h at 37°C in TE buffer (pH 8) with increasing concentrations of the [RuII(4-bptpy)(dmphen)Cl]ClO4. Lane 1, DNA control; lane 2, RuCl3 (100 µM); lanes 3–8, [RuII(4-bptpy)(dmphen)Cl]ClO4 complex: 25, 75, 125, 200, 300 and 400 µM, respectively.
![Figure 3. Agarose gel (1%) of pUC19 (100 µg/mL) incubated for 2 h at 37°C in TE buffer (pH 8) with increasing concentrations of the [RuII(4-bptpy)(dmphen)Cl]ClO4. Lane 1, DNA control; lane 2, RuCl3 (100 µM); lanes 3–8, [RuII(4-bptpy)(dmphen)Cl]ClO4 complex: 25, 75, 125, 200, 300 and 400 µM, respectively.](/cms/asset/d798c3dc-2fc0-4a57-88ba-48efab76a086/ienz_a_570007_f0004_b.gif)
Figure 4. The percentage of SC, OC and L forms of DNA produced by various concentration of [RuII(4-bptpy)(dmphen)Cl]ClO4 complex.
![Figure 4. The percentage of SC, OC and L forms of DNA produced by various concentration of [RuII(4-bptpy)(dmphen)Cl]ClO4 complex.](/cms/asset/a2add36a-586a-4277-a40e-9c5608f7f92a/ienz_a_570007_f0005_b.gif)
Time-dependent cleavage by complex 1 was studied at a concentration of 200 µM for 15–240 min (). As the incubation time increases, percentage of OC and L forms increases (). The linear form is produced only if incubated for ≥90 min. The SC form completely disappears at 240 min. So, the cleavage ability for all the complexes was checked at 200 µM concentration with incubation time of 240 min (). The amounts of linear DNA produced by complexes 1, 2 and 3 were 28%, 18% and 24%, respectively.
Figure 5. Agarose gel (1%) of pUC19 (100 µg/mL) at 37°C in TE buffer (pH 8) with 200 µM [RuII(4-bptpy)(dmphen)Cl]ClO4 complex for increasing reaction time. Lane 1, DNA control; lanes 2–8: 15, 30, 60, 90, 120, 180 and 240 min, respectively.
![Figure 5. Agarose gel (1%) of pUC19 (100 µg/mL) at 37°C in TE buffer (pH 8) with 200 µM [RuII(4-bptpy)(dmphen)Cl]ClO4 complex for increasing reaction time. Lane 1, DNA control; lanes 2–8: 15, 30, 60, 90, 120, 180 and 240 min, respectively.](/cms/asset/d49d4a0e-e734-4faf-94cd-762a055c16cc/ienz_a_570007_f0006_b.gif)
Conclusions
From the results of the DNA interaction studies, we were dragged to a conclusion that RuII complex with fluoro derivative of terpyridine shows lower DNA binding and cleavage activity. On the other hand, complex with bromo derivative of terpyridine shows higher DNA binding and cleavage activity. Complex 1 binds to DNA via classical intercalative mode. Complex 2 interacts with DNA electrostatically and complex 3 partially intercalates to DNA. The difference in Kb values may be due to different binding mode of the complexes.
Acknowledgement
The authors wish to thank Head, Department of Chemistry, and Dr. Thakkar, BRD school of Bioscience, Sardar Patel University, India for making it convenient to work in laboratory and U.G.C. for providing financial support under “UGC Research Fellowship in Science for Meritorious Students” scheme.
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
References
- Pyle A, Barton JK. Probing nucleic acids with transition metal complexes. Prog Inorg Chem 1990;38:413–475.
- Sigman DS, Mazumder A, Perrin DM. Chemical nucleases. Chem Rev 1993;93:2295–2316.
- Sundquist WI, Lippard SJ. The coordination chemistry of platinum anticancer drugs and related compounds with DNA. Coord Chem Rev 1990;100:293–322.
- Liu Y-J, Guan X-Y, Wei X-Y, He L-X, Mei W-J, Yao J-H. Ruthenium(II) complexes containing 2,9-dimethyl-1,10-phenanthroline and 4,4′-dimethyl-2,2′-bipyridine as ancillary ligands: synthesis, characterization and DNA-binding. Trans Met Chem 2008;33:289–294.
- Liu Y-J, Wei X-Y, Mei W-J, He L-X. Synthesis, characterization and DNA binding studies of ruthenium(II) complexes: [Ru(bpy)2(dtmi)]2+ and [Ru(bpy)2(dtni)]2+. Trans Met Chem 2007;32:762–768.
- Liu Y-J, Chen J-C, Wu F-H, Zheng K-C. Effects of substituent on the DNA-binding of ruthenium(II) complexes containing asymmetric tridentate intercalative ligands. Trans Met Chem 2009;34:297–305.
- Rajendiran V, Murali M, Suresh E, Palaniandavar M, Periasamy VS, Akbarsha MA. Non-covalent DNA binding and cytotoxicity of certain mixed-ligand ruthenium(II) complexes of 2,2′-dipyridylamine and diimines. Dalton Trans 2008;2157–2170.
- Vaidyanathan VG, Nair BU. Synthesis, characterization and DNA binding studies of a ruthenium(II) complex. j Inorg Biochem 2002;91:405–412.
- Harald H, Abdelkrim E, Albertus PHJ, Schuberta US. Synthesis and optical properties of (a)chiral terpyridine–ruthenium complexes. Tetrahedron 2004;60:6121–6128.
- Zhen QX, Zhang QL, Liu JG, Ye BH, Ji LN, Wang L. Synthesis, characterization and DNA binding of ruthenium(II) complexes containing the atatp ligand. j Inorg Biochem 2000;78:293–298.
- Liu J-G, Zhang Q-L, Ji L-N, Cao Y-Y, Shi X-F. Synthesis, characterization and interaction of mixed polypyridyl ruthenium(II) complexes with calf thymus DNA. Trans Met Chem 2001;26:733–738.
- Maheswari PU, Rajendiran V, Palaniandavar M, Parthasarathi R, Subramanian V. Synthesis, characterization and DNA-binding properties of rac-[Ru(5,6-dmp)2(dppz)]2+–enantiopreferential DNA binding and co-ligand promoted exciton coupling. J Inorg Biochem 2006;100:3–17.
- Tan L-F, Chao H. DNA-binding and photocleavage studies of mixed polypyridyl ruthenium(II) complexes with calf thymus DNA. Inorg Chim Acta 2007;360:2016–2022.
- Patel MN, Parmar PA, Gandhi DS. Square pyramidal copper(II) complexes with forth generation fluoroquinolone and neutral bidentate ligand: structure, antibacterial, SOD mimic and DNA-interaction studies. Bioorg Med Chem 2010;18:1227–1235.
- Adcock PA, Richard Keene F, Smythe RS, Snow MR. Oxidation of isopropylamine and related amines coordinated to ruthenium. Formation of monodentate imine and alkylideneamido complexes of ruthenium. Inorg Chem 1984;23:2336–2343.
- Trommel JS, Marzilli LG. Synthesis and DNA binding of novel water-soluble cationic methylcobalt porphyrins. Inorg Chem 2001;40:4374–4383.
- Mudasir S, Yoshioka N, Inoue H. DNA binding of iron(II) mixed-ligand complexes containing 1,10-phenanthroline and 4,7-diphenyl-1,10-phenanthroline. J Inorg Biochem 1999;77:239–247.
- Jin L, Yang P. Synthesis and DNA binding studies of cobalt (III) mixed-polypyridyl complex. J Inorg Biochem 1997;68:79–83.
- Zhang QL, Liu JG, Chao H, Xue GQ, Ji LN. DNA-binding and photocleavage studies of cobalt(III) polypyridyl complexes: [Co(phen)2IP]3+ and [Co(phen)2PIP]3+. J Inorg Biochem 2001;83:49–55.
- Chaires JB, Dattagupta N, Crothers DM. Studies on interaction of anthracycline antibiotics and deoxyribonucleic acid: equilibrium binding studies on interaction of daunomycin with deoxyribonucleic acid. Biochemistry 1982;21:3933–3940.
- Cohen G, Eisenberg H. Viscosity and sedimentation study of sonicated DNA-proflavine complexes. Biopolymers 1969;8:45–55.
- Yoshikawa N, Yamabe S, Kanehisa N, Kai Y, Takashima H, Tsukahara K. Syntheses, characterization, and DFT investigation of new mononuclear acetonitrile- and chloro-ruthenium(II) terpyridine complexes. Inorg Chim Acta 2006;359:4585–4593.
- Eva C, Hotze ACG, Tooke DM, Spek AL, Reedijk J. Ruthenium polypyridyl complexes containing the bischelating ligand 2,2′-azobispyridine. Synthesis, characterization and crystal structures. Inorg Chim Acta 2006;359:830–838.
- Goswami S, Chakravarty AR, Chakravorty A. Chemistry of ruthenium. 5. Reaction of trans-dihalobis[2-(arylazo)pyridine]ruthenium(II) with tertiary phosphines: chemical, spectroelectrochemical, and mechanistic characterization of geometrically isomerized substitution products. Inorg Chem 1982;21:2737–2742.
- Jiang CW, Chao H, Li H, Ji LN. Syntheses, characterization and DNA-binding studies of ruthenium(II) terpyridine complexes: [Ru(tpy)(PHBI)]2+ and [Ru(tpy)(PHNI)]2+. J Inorg Biochem 2003;93:247–255.
- Dalton SR, Glazier S, Leung B, Win S, Megatulski C, Burgmayer SJ. DNA binding by Ru(II)-bis(bipyridine)-pteridinyl complexes. J Biol Inorg Chem 2008;13:1133–1148.
- Nagababu P, Satyanarayana S. DNA binding and cleavage properties of certain ethylenediamine cobalt(III) complexes of modified 1,10-phenanthrolines. Polyhedron 2007;26:1686–1692.
- Yang G, Wu JZ, Wang L, Ji LN, Tian X. Study of the interaction between novel ruthenium(II)-polypyridyl complexes and calf thymus DNA. J Inorg Biochem 1997;66:141–144.
- Tan J, Wang B, Zhu L. DNA binding and oxidative DNA damage induced by a quercetin copper(II) complex: potential mechanism of its antitumor properties. J Biol Inorg Chem 2009;14:727–739.
![Figure 6. The percentage of SC, OC and L forms of DNA produced by 200 µM of [RuII(4-bptpy)(dmphen)Cl]ClO4 complex at different time.](/cms/asset/953c893c-f264-4d74-9e61-f4248512c5d9/ienz_a_570007_f0007_b.gif)
![Figure 7. Agarose gel (1%) of pUC19 (100 µg/mL) at 37°C in TE buffer (pH 8) with 200 µM compounds incubated for 4 h. Lane 1, DNA control; lane 2, RuCl3; lane 3, [RuII(4-bptpy)(dmphen)Cl]ClO4; lane 4, [RuII(4-fptpy)(dmphen)Cl]ClO4; lane 5, [RuII(4-mptpy)(dmphen)Cl]ClO4.](/cms/asset/53f93d64-968a-4654-9736-cf0216a61ebb/ienz_a_570007_f0008_b.gif)