Abstract
Herein, the compounds bearing sulfonamide fragment such as N-(2-amino-5-benzoylphenyl)-4-nitrobenzene sulfonamide hydrochloride (1), N-(quinolin-8-yl)-4-nitro-benzenesulfonamide hydrochloride (2), N-(pyridine-2-ylmethyl)-4-nitro-benzenesulfonamide hydrochloride (3) were synthesized by the reaction of 3,4-diaminobenzophenone, 8-aminoquinoline or 2-picoylamine and 4-nitrobenzensulfonyl chloride, respectively. The structures of the newly synthesized compounds were elucidated on the basis of elemental and spectral analyses. All the prepared compounds were evaluated for their in vitro anti-cancer activity against various cancer cell lines and to explore the underlying molecular mechanisms involved in this process. In vitro cytotoxic activities of the compounds were screened against human hepatocellular (HepG2), breast (MCF-7) and colon (Colo-205) cancer cell lines by MTT assay, mRNA expression of genes with qPCR and phosphorylation of p38 and ERK1/2 with Western blot. Tested compounds could significantly reduce cell proliferation and induced mRNA expression of pro-apoptotic genes; caspase 3, caspase 8 and caspase 9. Activation of these apoptotic genes probably is mediated by activation of p38.
Introduction
Cancer mortality rate in the developed world have risen throughout in spite of advancement made in cancer diagnosis and therapy during the last several decades and it is the leading cause of worldwide deathsCitation1. Previous observations suggested that induction of apoptosis contributed to the high rate of cell loss in malignant tumors and, moreover, could promote tumor progressionCitation2. Caspases are proteases, which have a central role in triggering and executing apoptosis. The two major pathways of triggering apoptosis are the intrinsic and extrinsic pathwayCitation3. The main intrinsic pathway is characterized by mitochondrial dysfunction, with the release of cytochrome c, activation of caspase-9, and subsequently of caspase-3. The extrinsic pathway is activated at the cell surface through death receptor mediated activation of caspase-8 or caspase-10, followed by caspase-3 activationCitation4.
Mitogen-activated protein kinases (MAPKs) play an essential role in apoptotic signal transduction by modulating gene transcription in the nucleus in response to changes in the cellular environmentCitation5,Citation6. The most studied MAPKs are the extracellular signal-regulated protein kinases (ERK1 and ERK2), and p38 MAPKs (p38α, p38β, p38γ, p38δ). Recently, it was observed that chemotherapeutic agents and other novel compounds initiate cancer cell apoptosis through the activation of p38 MAP kinaseCitation7.
ERK1/2 an inducible component of normal cellular signal transduction processes has been shown to be constitutively activated (phosphorylated) in several cancer cell linesCitation8–10. In contrast to such constitutive activity of ERK1/2, transient activation of the kinases may contribute to induction of apoptosisCitation11,Citation12. ERK activity has also been involved in cell death induced by various antitumor compounds, such as resveratrolCitation13, quercetinCitation14, miltefosineCitation15 and taxolCitation16.
The compounds bearing sulfonamide piece are generally occurred from R-sulfonyl chloride with primary or secondary amines in basic mediaCitation17. In recent times, there has been an increase in studies regarding the substances containing sulfonamide fragment because of their potential applications such as for coordination chemistryCitation18, medicinal chemistryCitation19, catalyst chemistryCitation20, chemical luminescenceCitation21 and analytical chemistry fieldsCitation22. On the other hand, sulfonamide compounds are known to contain inherent pharmacological properties such as anti-microbialCitation23, anti-inflammatoryCitation24, anti-viralCitation25, anti-proliferativeCitation26, angiogenesisCitation27 and enzyme inhibitory (carbonic anhydrase) activitiesCitation28.
Therefore, based on these findings, sulfonamide compounds were synthesized by the reaction of sulfonamide fragments; N-(2-amino-5-benzoylphenyl)-4-nitrobenzenesulfonamide hydrochloride (1), N-(quinolin-8-yl)-4-nitro-benzenesulfonamide hydrochloride (2), N-(pyridine-2-ylmethyl)-4-nitro-benzenesulfonamide hydrochloride (3) with3,4-diaminobenzophenone, 8-aminoquinoline or 2-picoylamine and 4-nitrobenzensulfonyl chloride, respectively and we tested the effects compounds on cell viability, pro-apoptotic gene expression and of p38/ERK activation.
Results and discussion
Synthesis
The aromatic sulfonamide compounds (1–3) were successfully synthesized by the reaction of aryl sulfonyl chloride with 3,4-diaminobenzophenone, 8-aminoquinoline or 2-picoylamine. In the 1H-NMR spectra for the synthesized compounds (1–3), for 1, the -H1, -H2, -H3 protons were located 7.08, 7.44 and 6.99 as doublet, respectively. the -Ha and -Hb could be were assigned 8.24 and 8.30 as doublet, respectively. For 2, the –Ha and –Hb protons were observed 8.11 and 8.30 as doublet, respectively. For 3, the -H1, -H2, -H3 and -H4 protons could be assigned 7.98 as doublet, 8.47 as triplet, 7.88 as triplet and 8.78 as doublet. The representative NMR spectrums are attached in Figure S1 as supplementary materials.
In the IR spectra for 1–3, N-H stretching frequency peaks belonging to the sulfonamide groups appeared at 3210, 3342 and 3239 cm−1 and NH2 stretching frequency peaks were observed at 3463–3380 for 1. In addition, the SO2 stretch peaks appeared at 1314–1155 for 1, 1350–1167 for 2, 1334–1160 for 3. All the measurements and interpretations are compatible with the proposed structure of compounds.
Biological activity
The sulfonamides constitute an important class of therapeutic agents in current medicinal science. Although they have a common chemical motif of an aromatic/heterocyclic sulfonamide, there are a variety of mechanisms for their anti-tumor action, such as disruption of microtubule assembly, cell cycle arrest in the G1 phase, angiogenesis inhibition and carbonic anhydrase inhibition. Furthermore, some of these compounds selected via elaborate preclinical screenings are currently being evaluated in clinical trialsCitation19.
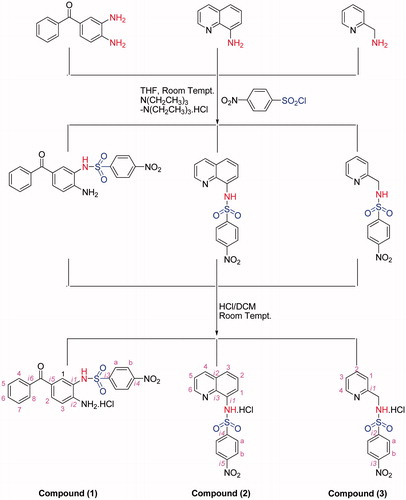
To test the effect of our compound, we used MTT assay which measure levels of cellular metabolism with decreased levels indicating cell death. Treatment with compounds during 24 h decreased cell viability in MCF-7 (), HEPG2 () and Colo-205 () cell lines. Decrease in cell viability in response to 12 and 24 h treatment with compounds correlated with a significant increase in pro-apoptotic gene expressions. Caspase 3 mRNA expression () increased following by 12 h treatment with our tested compounds in three different types of cancer cell lines. Especially in MCF-7 cells increasing in caspase 3 mRNA levels were measured after 24 h treatment with all tested compounds. Compound 1 and compound 2 treatments during 12 h might induce apoptosis via activating caspase 9 mRNA expressions in MCF-7 and HEPG2 cell lines (). In addition to these, Caspase 9 mRNA expression increased in MCF-7 cell line following 24 h treatments with compound 1 and compound 2. In both 12 and 24 h treatment periods, compound 2 caused to an increasing in caspase 8 mRNA expression in MCF-7 cell line ().
Figure 1. Proliferation inhibiting effects of compounds in MCF-7 cell line. n = 4, *p < 0.05 versus control.
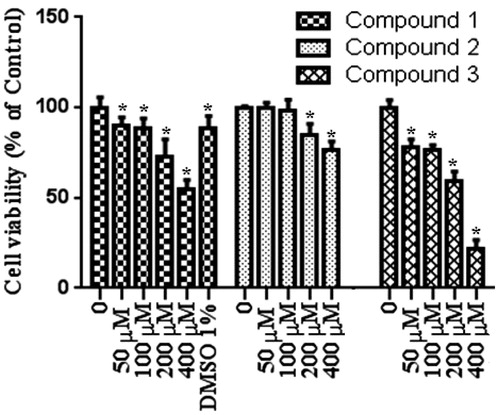
Figure 2. Proliferation inhibiting effects of compounds in HEPG2 cell line. n = 4, *p < 0.05 versus control.
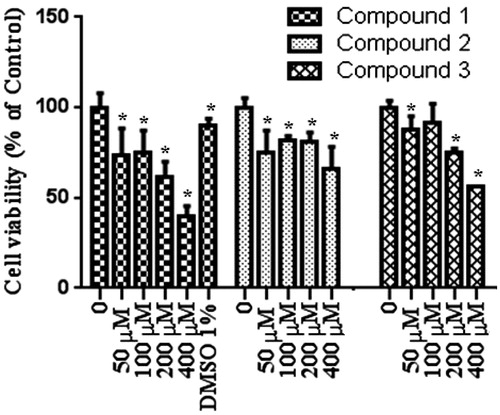
Figure 3. Proliferation inhibiting effects of compounds in Colo-205 cell line. n = 4, *p < 0.05 versus control.
Figure 4. (A) Caspase 3 mRNA levels following 12 h treatment with compounds, (B) Caspase 3 mRNA levels following 24 h treatment with compounds. Concentration of tested compounds were 400 μmol/L, n = 3, *p < 0.05 versus control.
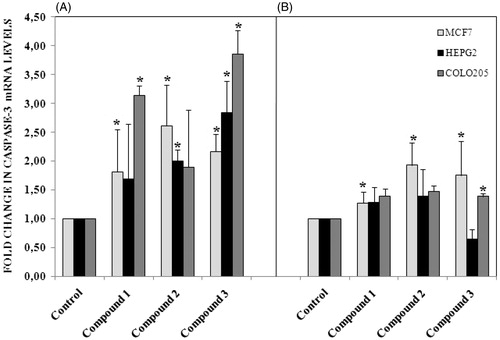
Figure 5. (A) Caspase 9 mRNA levels following 12 h treatment with compounds in different cancer cells, (B) 24 h treatment with compounds. n = 3, *p < 0.05 versus control.
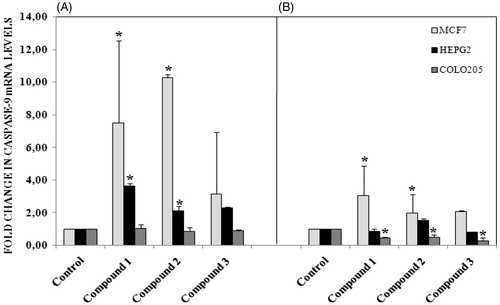
Figure 6. (A) Caspase 8 mRNA levels following 12 h treatment with compounds in different cancer cells, (B) 24 h treatment with compounds. n = 3, *p < 0.05 versus control.
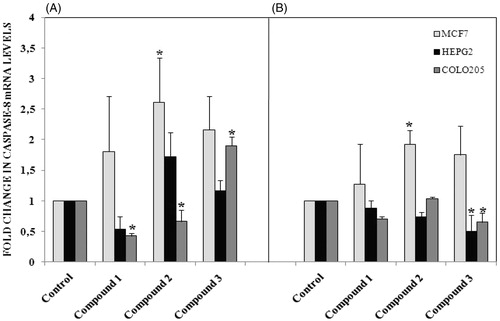
Based on the finding of apoptosis signaling activation in cancer cells, we were interested in which signaling pathway was involved in the activation of pro-apoptotic gene expression. Thus we further detected the activation of MAPK pathway. Therefore, activated ERK is usually translocated from the cytosol to the nucleus, where it can affect cellular functions, such as gene transcription, cell proliferation, differentiation, and apoptosisCitation20. Phosphorylation of ERK was markedly demonstrated in colo-205 cell line after 12 h treatment with all tested compounds (). Activation of caspase 3 mediated apoptosis in colo-205 cell line after 12 h treatments with all tested compounds might regulated with the activation of ERK phosphorylation. Activation of p38 MAPK was measured in HEPG2 and colo-205 cell lines following 12 h treatment with all tested compounds (). Interestingly, P38 phosphorylation decreased in 12 h treatment groups in MCF-7 cells but increased following 24 h treatments with all tested compounds.
Figure 7. (A) Activation of ERK phosphorylation following 12 h treatment with compounds in different cancer cells, (B) 24 h treatment with compounds. n = 3, *p < 0.05 versus control.
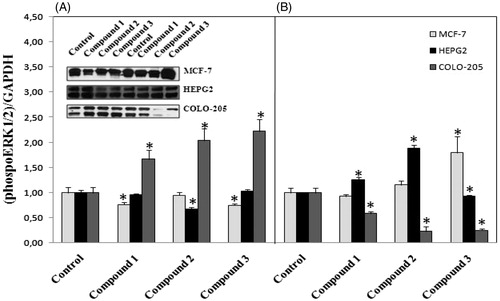
Figure 8. (A) Activation of P38 phosphorylation following 12 h treatment with compounds in different cancer cells, (B) 24 h treatment with compounds. n = 3, *p < 0.05 versus control.
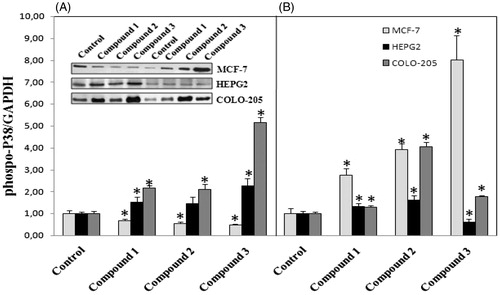
In conclusion, all tested compounds had anti-proliferative effects in different cancer cell lines. Decrease in cell viability in response to 12 and 24 h treatment with compounds correlated with a significant increase in pro-apoptotic gene expressions. The underlying molecular mechanisms involved in apoptotic cancer cell death after treatment with our newly synthesized compounds might be the activation of MAPK pathways.
Experimental
All chemicals used in the synthesis process (reagents and solvents) were purchased from chemical companies (Sigma-Aldrich, Merck and Alfa Aesar) and used as received unless otherwise stated. The 400 MHz 1H-NMR and 100.56 MHz 13C-NMR spectra were recorded at ambient temperature on a Bruker 400 NMR spectrometer in CDCl3 as solvent. NMR signals are given in parts per million (ppm) as δ downfield from tetramethylsilane (TMS) (δ 0.00) as an internal standard. Coupling constants are given in Hertz. The multiplicity of NMR peaks is abbreviated as follows: br = broad, d = doublet, m = multiplet, s = singlet, t = triplet. The elemental analyses were carried out using a Truspec MICRO (LECO) instrument. A Perkin-Elmer Spectrum 400 FTIR system with universal ATR sampling accessory was used to obtain the FT-IR spectra. For melting point determination, an Electrothermal 9100 instrument was used with open capillary tubes in Department of Chemistry, Faculty of Science, Erciyes University, Kayseri, Turkey.
General procedure for the synthesis of aromatic sulfonamide compounds 1–3
Stage-1
A solution of N(CH2CH3)3 (15 mmol) in THF (tetrahydrofuran) (5 ml) was slowly added to a solution of R-sulfonyl chlorides (10 mmol) in THF (10 ml), immediately followed by 3,4-diaminobenzophenone, 8-aminoquinoline or 2-picoylamine, (10 mmol) in THF (5 ml). After the desired time, the reaction mixture was filtered and the volatiles were removed in vacuo. The residue was dissolved in DCM (dichloromethane) (20 ml) and washed with H2O (3 × 10 ml), dried over anhydrous MgSO4, filtered and concentrated to half of its volume under reduced pressure. The solution was purified by precipitation with diethyl ether from a CH2Cl2 solution to give a microcrystalline product. All the desired products were checked by FT-IR and NMR.
Stage-2
The HCl solution (5 mmol) was slowly added to DCM solutions of the each desired products (5 mmol) with a dropping funnel and stirred at ambient temperature in a Schlenk tube for 1 h. After the desired time, the mixture was filtered. The crude product can be used without further purification. An analytically pure sample can be characterized with NMR, FT-IR and EA (Scheme 1).
Data for the compounds 1–3
For N-(2-amino-5-benzoylphenyl)-4-nitrobenzenesulfonamide hydrochloride (1)
Color: Light yellow. Yield: 76%. M.P.: 202 °C. 1H-NMR (CDCl3, δ ppm): 6.89 (d, 1H, J = 8 Hz, -H3), 7.08 (s, 1H, -H1), 7.44 (d, 1H, J = 8 Hz, -H2), 7.54–7.89 (m, 5H, -H4,5,6,7,8), 8.24 (d, 2H, J = 8 Hz, -Ha), 8.41 (d, 2H, J = 8 Hz, -Hb). 13C-NMR (CDCl3, ppm): 113.5 (-C1), 117.0 (-C3), 124.4 (-Cb), 128.3 (-Ca), 129.2 (-C5,7), 130.4 (-C4,8), 131.8 (-Ci1), 132.1 (-Ci5), 133.2 (-C6), 135.8 (-Ci6), 143.9 (-Ci2), 150.3, (-Ci3), 151.1 (-Ci4), 193.5 (-C=O).IR (cm−1): 3463 (NH2), 3380 (NH2), 3210 (-NH), 3061, 2986, 2770, 2532, 1662 (-C=O), 1625, 1597, 1577, 1533, 1507, 1484, 1405, 1374, 1354, 1314 (νas-SO2), 1303, 1285, 1238, 1181, 1155 (νs-SO2), 1126, 1109, 1081, 1047, 999, 975, 941, 909, 892, 872, 854, 841, 826, 795, 737, 722, 698, 680, 626, 608, 589, 568, 540, 453. Anal. Calcd. for: [C19H16ClN3O5S], C: 52.60, H: 3.72, N: 9.68, S: 7.39. Found: C: 52.82, H: 3.91, N: 9.49, S: 7.74.
For N-(quinolin-8-yl)-4-nitro-benzenesulfonamide hydrochloride (2)
Color: Light yellow. Yield: 69%. M.P.: 216 °C. 1H-NMR (CDCl3, δ ppm): 6.07 (-NH), 7.57–7.87, (m, 4H, -H1,2,3,5), 8.11 (d, 2H, J = 8 Hz, -Ha), 8.30 (d, 2H, J = 8 Hz, -Hb), 8.57 (d, 1H, J = 8 Hz, -H4), 8.92 (d, 1H, J = 8 Hz, -H6). 13C-NMR (CDCl3, ppm): 122.1 (-C3), 122.9 (-C5), 124.8 (-Cb), 125.6 (-C1), 127.1 (-C2), 129.0 (-Ca), 129.1 (-Ci2), 132.4 (-C4), 135.7 (-Ci3), 139.2 (-Ci1), 145.4 (-Ci4), 149.4 (-C6), 150.3 (-Ci5). IR (cm−1): 3342 (NH), 3096, 2958, 2806, 2741, 1634, 1606, 1593, 1557, 1532, 1469, 1423, 1403, 1368, 1350 (νas-SO2), 1301, 1257, 1215, 1167 (νs-SO2), 1088, 1038, 1014, 947, 916, 878, 854, 838, 809, 797, 776, 761, 738, 682, 637, 611, 601, 560, 528, 483, 465. Anal. Calcd. for: [C15H12ClN3O4S], C: 49.25, H: 3.31, N: 11.49, S: 8.77. Found: C: 49.51, H: 3.19, N: 11.61, S: 8.93.
For N-(pyridine-2-ylmethyl)-4-nitro-benzenesulfonamide hydrochloride (3)
Color: Light yellow. Yield: 73%. M.P.: 224 °C. 1H-NMR (CDCl3, δ ppm): 4.53 (d, 2H, J = 4 Hz, -CH2-NH-) 7.88 (t, 1H, J = 8 Hz, -H3), 7.98 (d, 1H, J = 8 Hz, -H1), 8.12 (d, 2H, J = 8 Hz, -Ha), 8.38 (d, 2H, J = 8 Hz, -Hb), 8.47 (t, 1H, J = 8 Hz, -H2), 8.78 (d, 1H, J = 8 Hz, -H4), 10.61 (-NH).13C-NMR (CDCl3, ppm): 49.0 (-CH2-NH-) 113.3 (-C1), 116.16 (-C3), 125.1 (-Cb), 128.9 (-Ca), 142.3 (-C2), 145.6 (-Ci2), 145.8 (-C4), 150.2 (-Ci3), 153.6 (-Ci1). IR (cm−1): 3239 (NH), 3102, 3068, 2853, 2390, 2051, 1987, 1939, 1616, 1536, 1473, 1431, 1399, 1362, 1334 (νas-SO2), 1311, 1290, 1233, 1180, 1160 (νs-SO2), 1093, 1036, 1007, 964, 852, 823, 771, 749, 737, 679, 648, 626, 604, 557, 532, 515, 462. Anal. Calcd. for: [C12H12ClN3O4S], C: 43.71, H: 3.67, N: 12.74, S: 9.72. Found: C: 43.82, H: 3.53, N: 12.55, S: 9.81.
Cell culture
The cell lines (Human colon carcinoma cell line Colo-205, Human hepatocellular carcinoma cell line HepG2 and Human breast adenocarcinoma cell line MCF7, Human Lung adenocarcinomic alveolar basal epithelial cell line A549) used in the study were purchased from American Type Culture Collection (ATCC). Colo-205 were maintained in RPMI 1640 Medium and HepG2 and MCF7 cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM). Cell cultures were supplemented with 10% fetal bovine serum (FBS), 2 mM l-Glutamine, 10 000 units/ml penicillin, and 10 mg/mL streptomycin and maintained under an atmosphere of 5% CO2 with 90% relative humidity at 37 °C, according to standard protocols.
MTT reduction assay
Anti-proliferative effect of the compounds (1–3) was evaluated by MTT reduction assay. Cells were treated with 0–400 µM of compounds (compound 1 and 2 dissolved in DMSO 5% and 2.5 %, respectively). Compound 3 dissolved in water. Final concentration of DMSO 1% for compound 1 treatment and 0.5% for compound 2 treatment in wells) in without phenol red 10% FBS supplemented mediums for 24 h and then rinsed 3 times with ice cold PBS. MTT was added to the final concentration of 0.5 mg/mL. After from MTT incubation (4 h), solubilization buffer (10% sodium dodecyl sulfate in 0.01 mol/L HCl) was added and the colored formazan crystals were gently re-suspended. The absorbance data at 570 nm were recorded with a microplate reader (Bio-Tek ELX800, BioTek Instruments Inc., Winooski, VT).
Quantitative real-time PCR
Total RNA isolation from cells was performed via phenol-guanidine thiocyanate extraction using TriPure isolation reagent (Roche Diagnostics GmbH, Mannheim, Germany), according to the manufacturer’s instructions. Total RNA (1 µg) was reverse-transcribed to cDNA using a Transcriptor High Fidelity cDNA Synthesis Kit (Roche Diagnostics GmbH, Mannheim, Germany) in a 20 µL reaction mixture. Real-time PCR was carried out using a Light Cycler Nano System (Roche Diagnostics GmbH, Mannheim, Germany). To quantify cDNA, qPCR was performed using Fast Start Essential DNA probe master mix (Roche Diagnostics GmbH, Mannheim, Germany). The reaction mixture (15 µL) was prepared in Ligh Cycler 8-tube strips (Roche Diagnostics GmbH, Mannheim, Germany) and included 10 µL of 2× Master Reaction Mix (Roche Diagnostics GmbH, Mannheim, Germany), 4 µL PCR grade water, 1 µL catalogue assay kit (Caspase9, Caspase 8, Caspase 3, β-actin) and 5 µL of cDNA. Real-time PCR was performed according to following conditions: activation of Taq DNA polymerase and DNA denaturation at 95 °C for 10 min, followed by 45 amplification cycles for 10 s at 95 °C and for 30 s at 60 °C. For each sample the level of target gene transcripts was normalized to β-actin.
Western blot analysis
Cells were incubated with 400 μM concentrations of compounds for 12 h and 24 h. Cellular protein extract (20–25 μg) isolated with RIPA buffer (Sigma-Aldrich, St. Louis, MO) supplemented with protease and phospatase inhibitory cocktail (Santa Cruz Biotechnology, Santa Cruz, CA), was subjected to discontinuous 5% to 12% SDS-PAGE. The primary antibodies were used rabbit-anti-phospo-p38 (1:1000 Cell signaling, Danvers, MA). After serial washes, primary antibodies were detected by secondary antibodies conjugated with horseradish peroxidese. Specific signals were visualized with enhanced chemiluminescence detection kits (Pierce WestPico, Rockford, IL).
Statistical analysis
Data are expressed as mean ± standard deviation (SD). Significance of the difference between the control and treated group(s) was assessed by Student’s t-test. Values of p < 0.05 were taken to be significant. Expression of the target genes’ mRNA corresponding to that of the housekeeping gene mRNA was analyzed by a relative expression software tool (REST 2009, v.2.0.13). The statistical significance was established via the pair-wise fixed reallocation randomization test. p Values <0.05 were reflected on statistically significant.
Conclusion
Herein, we synthesized the new compounds (1–3) bearing sulfonamide moiety and tested cytotoxic effects of compounds (more than 3 compounds (data unpublished)) but, only three compound (Compound 1–3) were active (decreased cancer viable cell number) according to the results of MTT assay. Lead compounds 1, 2, and 3 merit further studies (qPCR and western blot analysis) as novel promising anti-cancer agents against different cell lines.
Furthermore, all the experiments show that to us, (1) compound was generally found to be the most active compound compared to others for cells viability. So, the 3,4-diaminobenzophenonesulfonamide derivative is superior from others and this compound may be derivatized for anti-cancer activity in the future. In addition, it is clear that the N-(2-amino-5-benzoylphenyl)-4-nitrobenzenesulfonamide hydrochloride (1), N-(quinolin-8-yl)-4-nitro-benzenesulfonamide hydrochloride (2) and N-(pyridine-2-ylmethyl)-4-nitro-benzenesulfonamide hydrochloride (3) compounds could significantly reduce cell proliferation and induced mRNA expression of pro-apoptotic genes; caspase 3, caspase 8 and caspase 9. The fabricated compounds bearing sulfonamide moiety can be used herein show moderate efficiency as anti-cancer agents for treatment of various malignanciesCitation30.
Supplementary material available online.
Supplementary Figure S1.
Supplemental Material.pdf
Download PDF (117.2 KB)Declaration of interest
The authors declare that they have no conflict of interest.
References
- Thun MJ, DeLancey JO, Center MM, et al. The global burden of cancer: priorities for prevention. Carcinogenesis 2010;1:100–10
- Johnstone RW, Ruefli AA, Lowe SW. Apoptosis: a link between cancer genetics and chemotherapy. Cell 2002;108:153–64
- Cohen GM. Caspases: the executioners of apoptosis. Biochem J 1997;326:1–16
- Hengartner MO. The biochemistry of apoptosis. Nature 2000;407:770–6
- Leelahavanichkul K, Amornphimoltham P, Molinolo AA, et al. A role for p38 MAPK in head and neck cancer cell growth and tumor-induced angiogenesis and lymphangiogenesis. Mol Oncol 2014;1:105–18
- Cuenda A, Rousseau S. p38 MAP-kinases pathway regulation, function and role in human diseases. Biochim Biophys Acta 2007;1773:1358–75
- Lenassi M, Plemenitaš A. The role of p38 MAP kinase in cancer cell apoptosis. Radiol Oncol 2006;40:51–6
- Ostrowski J, Trzeciak L, Kolodziejski J, Bomsztyk K. Increased constitutive activity of mitogen-activated protein kinase and renaturable 85 kDa kinase in human-colorectal cancer. Brit J Cancer 1998;78:1301–6
- Kim SC, Hahn JS, Min YH, et al. Constitutive activation of extracellular signal-regulated kinase in human acute leukemias: combined role of activation of MEK, hyperexpression of extracellular signalregulated kinase, and down regulation of a phosphatase, PAC1. Blood 1999;93:3893–9
- Chen Z, Sun J, Pradines A, Favre G, et al. Both farnesylated and geranylgeranylated RhoB inhibit malignant transformation and suppress human tumor growth in nude mice. J Biol Chem 2000;275:17974–8
- Ishikawa Y, Kitamura M. Dual potential of extracellular signal-regulated kinase for the control of cell survival. Biochem Biophys Res Commun 1999;264:696–701
- Pavlovic D, Andersen NA, Mandrup-Poulsen T, Zizirik DL. Activation of extracellular signal-regulated kinase (ERK)1/2 contributes to cytokineinduced apoptosis in purified rat pancreatic beta-cells. Eur Cytokine Netw 2000;11:267–74
- Shih A, Davis FB, Lin HY, Davis PJ. Resveratrol induces apoptosis in thyroid cancer cell lines via a MAPK- and p53-dependent mechanism. J Clin Endocrinol Metab 2002;87:1223–32
- Kim YH, Lee DH, Jeong JH, et al. Quercetin augments TRAIL-induced apoptotic death: involvement of the ERK signal transduction pathway. Biochem Pharmacol 2008;75:1946–58
- Tewari R, Sharma V, Koul N, Sen E. Involvement of miltefosine-mediated ERK activation in glioma cell apoptosis through Fas regulation. J Neurochem 2008;107:616–27
- Bacus SS, Gudkov AV, Lowe M, et al. Taxol-induced apoptosis depends on MAP kinase pathways (ERK and p38) and is independent of p53. Oncogene 2001;20:147–55
- Jones DN. Comprehensive organic chemistry. Vol. 3. Oxford: Pergamon Press; 1979:345
- González-Alvarez M, Alzuet G, Borrás J, et al. Comparison of protective effects against reactive oxygen species of mononuclear and dinuclear Cu(II) complexes with N-substituted benzothiazolesulfonamides. Inorg Chem 2005;44:9424–33
- Shah SS, Rivera G, Ashfaq M. Recent advances in medicinal chemistry of sulfonamides. Rational design as anti-tumoral, anti-bacterial and anti-inflammatory agents. Mini Rev Med Chem 2013;13:70–86
- Dayan S, Kalaycıoğlu NÖ, Dayan O, et al. Synthesis and characterization of SiO2-supported ruthenium complexes containing aromatic sulfonamides: as catalysts for transfer hydrogenation of acetophenone. Dalton Trans 2013;14:4957–69
- Turkmen H, Zengin G, Buyukkircali B. Synthesis of sulfanilamide derivatives and investigation of in vitro inhibitory activities and antimicrobial and physical properties. Bioorg Chem 2011;3:114–19
- Chohan ZH, Shad HA, Supuran CT. Synthesis, characterization and biological studies of sulfonamide Schiff's bases and some of their metal derivatives. J Enzyme Inhib Med Chem 2012;1:58–68
- Gangapuram M, Mazzio E, Eyunni S, et al. Synthesis and biological evaluation of substituted N-[3-(1H-Pyrrol-1-yl)methyl]-1,2,5,6-tetrahydropyridin-1-yl]benzamide/benzene sulfonamides as anti-inflammatory agents. Arch Pharm (Weinheim) 2014;5:360–9
- Chen Z, Xu W, Liu K, et al. Synthesis and antiviral activity of 5-(4-chlorophenyl)-1,3,4-thiadiazole sulfonamides. Molecules 2010;12:9046–56
- Bashandy MS, Alsaid MS, Arafa RK, Ghorab MM. Design, synthesis and molecular docking of novel N,N-dimethylbenzenesulfonamide derivatives as potential antiproliferative agents. J Enzyme Inhib Med Chem 2013;3:1–9
- Funahashi Y, Sugi NH, Semba T, et al. Sulfonamide derivative, E7820, is a unique angiogenesis inhibitor suppressing an expression of integrin alpha2 subunit on endothelium. Cancer Res 2002;21:6116–23
- Güzel O, Innocenti A, Vullo D, et al. 3-phenyl-1H-indole-5-sulfonamides: structure-based drug design of a promising class of carbonic anhydrase inhibitors. Curr Pharm Des 2010;29:3317–26
- Owa T, Okauchi T, Yoshimatsu K, et al. A focused compound library of novel N-(7-indolyl)benzenesulfonamides for the discovery of potent cell cycle inhibitors. Bioorg Med Chem Lett 2000;11:1223–6
- McCubrey JA, Steelman LS, Chappell WH, et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim Biophys Acta 2007;8:1263–84
- Ghorab MM, El-Gazzar MG, Alsaid MS. Synthesis and anti-breast cancer evaluation of Novel N-(Guanidinyl)benzenesulfonamides. Int J Mol Sci 2014;15:5582–95