Abstract
Withaferin A is an abundant withanolide present in Withania somnifera leaves and to some extent in roots. It has been known for its profound anti-cancer properties, but its role in counteracting the Leishmania donovani infection has to be explored. Pteridine reductase 1 (PTR1) is involved in pteridine salvage and an important enzyme for the parasite growth, which could be targeted for the development of an efficient antileishmanial drug. We employed molecular docking studies to identify the binding mode of withaferin A with PTR1 in silico. We further cloned, expressed, and purified PTR1 of L. donovani and performed the enzyme kinetics using the Michaelis–Menten equation and enzyme inhibition studies with withaferin A by plotting the Lineweaver–Burk graph, which followed an uncompetitive mode of inhibition. We also showed the inhibition of the enzyme in the crude lysate of treated parasites. Thus, our study contributes towards understanding the mode of action of withaferin A against L. donovani parasite.
Introduction
According to the WHO statistics, 1.5–2 million new leishmaniasis cases are being reported annually which affects 12 million people living in 88 countriesCitation1. This disease is more prevalent in low-income groups residing in the endemic region, out of which 50% of the visceral leishmaniasis (VL) cases are from the Indian subcontinentCitation2. Current chemotherapy for leishmaniasis includes pentavalent antimonials, amphotericin B, and more recently introduced first oral drug miltefosine. But the factors like emergence of resistance, severe side effects, high cost, and low efficacy have been hindering the usage of these drugs Citation3–5. The failure of these drugs in chemotherapy necessitates for the development of alternative drugs. In this regard, our previous study showed the effective antileishmanial activity of withanolides through apoptotic like death mechanismCitation6 and withaferin A is an abundant compound among the withanolides in the W. somnifera leaves. Therefore, understanding the novel biochemical pathways in the parasite will pave path for developing selective antileishmanial drugs. In this regard, we chose PTR 1 as the target enzyme, which is exclusively present in Leishmania parasites and is very essential for the growth of the parasites Citation7,Citation8.
Leishmania and other trypanosomatid protozoans are auxotrophs for reduced pteridines (pterins and folates), which are required for critical cellular pathways like nucleic acid and protein biosynthesis. The predominant role of PTR1 is to salvage oxidized pterins rather than to reduce folates and it is the only enzyme that has been reported to reduce biopterin for the in vivo growth of Leishmania Citation8–11. Thus, pterin compounds like biopterin or folate are acquired from the host and active tetrahydro-species are generated by successive reductions of pterin compounds that are carried by two bifunctional enzymes namely dihydrofolate reductase-thymidylate synthase (DHFR-TS) and pteridine reductase (PTR1). The former reduces folate and 7,8-dihydrofolate (DHF) to 5,6,7,8-tetrahydrofolate (THF)Citation7 and the latter catalyzes NADPH-dependent reversible reduction of oxidized pterins to dihydrobiopterin (DHB) as well as of tetra-hydrobiopterin (THB) and folates to DHF and THFCitation7. Although it catalyzes the same reaction as that of DHFR, the failure of anti-folate strategies lies in the PTR1 resistance to DHFR inhibitors like methotrexate. The intriguing feature of PTR1 makes it a suitable drug target for the development of antileishmanial agents Citation7,Citation12. In this regard, PTR1 presents an attractive drug target for the development of antileishmanial agents.
Withania somnifera is also known as Ashwagandha or Indian ginseng in the Ayurvedic medical systems. It has been used as an indigenous herb for more than 3000 yearsCitation13. The pharmacological activities of this plant have been attributed to its secondary metabolites called withanolidesCitation14. Withanolides are C28-steroidal lactones with the intact or rearranged ergostane framework. Withaferin A is one of the most predominant compounds among the withanolides. Previous studies have reported various biological activities of the crude rootCitation15 and leaf extractsCitation16–18 of W. somnifera. Withaferin A, an abundant withanolide, has potent anticancer and anti-proliferative activities against a wide range of cancer cell types Citation19–21. Our previous study showed that withanolides from W. somnifera have been able to induce apoptotic like death in L. donovani promastigotes; however, the molecular mechanism of these compounds has yet to be deciphered in both stages of the parasiteCitation6.
In our present study, we showed the antileishmanial activity of withaferin A on L. donovani promastigotes and also an attempt was made to find the mode of action from in silico analysis by retrieving the 3D structure of L. donovani. The docking studies of withaferin A and PTR1 enzyme gave an insight into the mechanism of action of withaferin A on the L. donovani parasites. Subsequently, LdPTR1 was cloned, expressed, and purified in native form. The purified recombinant enzyme was tested for enzyme activity by using the Michaelis–Menten equation and kinetics of inhibition was also studies using the Lineweaver–Burk plot. Interestingly, our results proved that withaferin A indeed inhibit the PTR1 enzyme from withaferin A treated parasites. Thus, the current study validates the PTR1 enzyme as a target of withaferin A thus providing a hint to decipher the antileishmanial mechanism of withaferin A.
Methods
Materials
Withaferin A was purchased from Sigma Aldrich Chemicals, Delhi, India. The compound was dissolved in methanol (HPLC Grade, Qualigens) at a concentration of 1 mg/ml. It was diluted with M199 media prior to use. Leishmania donovani strain DD8 (MH0M/IN/80/DD8) was procured from ATTC, Elizabeth City, NC. The parasites were cultured in vitro in M199 medium supplemented with 15% FBS, 100 U/ml of penicillin and 100 mg/ml of streptomycin at 25 °C.
Antileishmanial effect of withaferin A in vitro
To assess the antileishmanial activity of withaferin A against L. donovani parasites, MTT assay was performed. Briefly, exponentially growing L. donovani (1 × 106) parasites were treated with varied concentrations of withaferin A (1–25 μM) in triplicates for 48 h at 25 °C. MTT reagent was added to the wells at the concentration of 100 μg/ml and incubated at 37 °C for 4 hrs. The formazan crystals were dissolved by adding 100 μl DMSO. The absorbance was read at 540 nm and results were expressed as the percentage of viability.
Docking analysis
Python 2.7, Cygwin c:\program, Python 2.5, Molecular Graphics Laboratory (MGL, New York, NY), AutoDock4.2 and Discovery Studio Visualizer 2.5.5 were downloaded from online sources.
The docking of withaferin A into the binding pocket of PTR1 protein was performed using Autodock software (The Scripps Research Institute, La Jolla, CA), a powerful tool for molecular recognition. To validate the molecular modeling programs, we evaluated the docking accuracies of Autodock by docking withaferin A into the binding site.
Protein preparation for docking
The 3D structure of Leishmania donovani PTR1 (PDB ID: 2XOX) was retrieved from Protein Data Bank (PDB) (http://www.pdb.org/pdb/home/home.do). Before the initiation of docking simulations, all non-protein molecules were removed from PTR1; for any alternative atom locations, only the first location was retained. All the docking calculations were performed using Autodock 4.2 (The Scripps Research Institute, La Jolla, CA). PTR1 was modified by adding polar hydrogens and then kept rigid in the docking process, whereas Ligand Module in Autodock Tools set all the torsional bonds of ligands free.
Ligand preparation for docking
The ligand withaferin A was built using Chemsketch and optimized using “Prepare Ligands” in the AutoDock 4.2 for docking studies. The optimized ligand molecules were docked into refined PTR1 model using “LigandFit” in the AutoDock 4.2.
Binding sites of these complexes were identified as described previously. The conformation of the binding site was constructed manually to accommodate withaferin A. The validation of the docking accuracy was done by docking withaferin A into the binding site of PTR1. AutoDock binding affinities of withaferin A into PTR1 were evaluated by the binding free energies (kcal/mol), inhibition constants (Ki), hydrogen bonds, and RMSD values.
Cloning, expression, and purification of PTR1
DNA isolation
For nuclear DNA isolation, 10–15 ml of promastigote culture was taken and parasites were harvested at 5000 rpm for 8 min at 4 °C. The supernatant was decanted; pellet was washed with 3–6 ml NET buffer (0.1 M NaCl, 1.0 mM EDTA pH 8.0, and 10 mM Tris pH 8.0) and centrifuged at 5000 rpm for 8 min at 4 °C. The supernatant was discarded and the pellet was resuspended in 750 μl NET buffer, 7.5 μl proteinase K (10 mg/ml stock), and 50 μl of 15% sarcosyl. The sample was incubated at 37 °C overnight for proteinase K activity. The cell lysate was centrifuged at 18 000 rpm for 1 h at 4 °C. The supernatant containing nuclear DNA was transferred to a fresh tube and treated with RNase at 37 °C for 1 h. DNA was extracted first with one volume phenol (750 μl), then with phenol/chloroform/isoamyl alcohol (25:24:1) and finally with chloroform. DNA was precipitated with two volumes of prechilled absolute ethanol, washed twice with 70% ethanol, and the pellet was stored at 4 °C for future use.
Cloning and expression of PTR1
PCR amplification was carried out using Phusion High-Fidelity DNA polymerase (Thermo Scientific, Waltham, MA). Reactions were carried out in a Veriti 96 well thermal cycler (Applied Biosystems, Hempstead, UK) with nuclear DNA as a template. A set of primers were designed from L. donovani PTR1 sequence (GenBank accession no. XM_003860884). The sequence of forward primer includes EcoRI restriction site (underlined) 5′-ATTGGAATTCATGGCTGCTCCGACC-3′ and the reverse primer includes XhoI restriction site (underlined) 5′-AATTCTCGAGTCAGGCCCGGGTAA-3′. The PTR1 coding region was amplified using the genomic DNA as a template. The PCR reaction was carried out using 35 cycles of 10 s at 98 °C, 30 s at 56 °C, and 30 s at 72 °C. The reaction mixture was incubated for 30 s at 98 °C and 7 min at 72 °C before and after PCR cycling, respectively. The PCR product was separated by electrophoresis on a 1% agarose gel and then stained with ethidium bromide. The DNA band was visualized under ultraviolet light (UV trans-illuminator).
The amplified PTR1 PCR product was extracted using the gel extraction kit (Machery-Nagel, Deer Park, NY). The eluted PCR product and pET28a vector were double digested with EcoRI and XhoI and ligated with T4 DNA ligase. The recombinant plasmid was transformed into E. coli BL21 (DE3) cells and positive colonies were selected using the kanamycin resistance marker. The PTR1 protein expression was induced in logarithmic phase culture (at OD600 of ∼0.5–0.6) for 4 h with 1 mM IPTG. Both the uninduced and induced cells were harvested and lysed in 2 × sample buffer (100 mM Tris-HCl pH-8, 20% glycerol, 4% SDS, 2% 2-ME, and 0.2% bromophenol blue) and separated on 12% SDS-PAGE.
SDS–polyacrylamide gel electrophoresis (SDS–PAGE) and western blotting
Uninduced and induced cell lysates were separated by 12% SDS–PAGE and transferred onto a nitrocellulose membrane (Pall Life Sciences, Washington, NY). The membrane was blocked with blocking buffer (5% (w/v) skimmed milk powder in 1 × TBS-0.1% Tween20) for 3 h at room temperature. Subsequently, it was incubated for 1 h with primary anti-His antibody (1:2000) in 1 × blocking buffer. Later, the blot was washed thrice with 1 × TBS-T (10 min each) and incubated for 1 h with secondary antibody conjugated to ALP (Rabbit anti-mouse IgG (Sigma, St. Louis, MO), 1:10 000) diluted in blocking buffer followed by three washes with 1 × TBS-T. Finally, the membrane was developed with BCPIP/NBT premix (Sigma, St. Louis, MO) substrate for the visualization of bands.
Purification of PTR1 protein
The harvested induced cell pellet was resuspended in 5 ml lysis buffer (50 mM Tris pH 8.0, 0.5 M NaCl, 5 mM2-ME, 1% Triton X-100, and 10% glycerol) and sonicated on ice at 35 W with 5 pulses for 30 s each. The resulting cell lysate was centrifuged at 14 000 rpm for 20 min at 4 °C and the supernatant (soluble fraction) containing the recombinant protein with N-terminal His6 tag was subjected to affinity chromatography using Ni-NTA column (GE Healthcare, Buckinghamshire, UK). Briefly, the supernatant collected was applied onto the column and allowed to bind for 2 h at 4 °C. The column was washed thrice with wash buffer (50 mM Tris pH8.0, 0.5 M NaCl, and 20 mM imidazole) and then the bound protein was eluted with elution buffer (50 mM Tris pH7.5, 0.5 M NaCl, and 200 mM imidazole). The eluted protein was pooled and dialyzed overnight at 4 °C against the dialysis buffer (25 mM Tris pH 7.5, 100 mM NaCl, and 10% glycerol) with an intermittent change of buffer at 6 h interval.
PTR1 enzyme activity and in vitro inhibition assay of the recombinant enzyme (LdPTR1)
Reductase activity of PTR1 was assayed and the volume of each reaction was 500 μl. The assay was initiated by the addition of substrate after incubation at 30 °C for 45 min at the zero time point, and the rates were measured for 3 min with 1 min intervals. NADPH (HiMedia, Mumbai, India) oxidation was monitored at 340 nm. The enzyme assay for LdPTR1 were performed at 30 °C in the presence of NADPH (100 μM) and varied concentrations (0–120 μM) of substrate biopterin (Sigma, St. Louis, MO) in 20 mM sodium acetate buffer pH 4.8 (Sigma, St. Louis, MO). Since pteridines exhibit absorbance changes when reduced, the extinction coefficient 7,230 M−1 cm−1 was used for the coupled oxidation/reduction of NADPH/biopterinCitation8. The kinetic parameters Km and Vmax for the Pteridine substrates were evaluated by fitting the Michaelis–Menten equation by non-linear regression. For inhibition studies, recombinant LdPTR1 was incubated with withaferin A and NADPH and the reaction initiated with the substrate biopterin. Lineweaver–Burk plots were used to determine the mode of inhibition by an inhibitor, examining ligand competition under conditions where [inhibitor] ≪ [enzyme] and [biopterin].
PTR1 enzyme assay in treated crude lysates of L. donovani parasites
Leishmania donovani promastigotes (1 × 106) were treated with withaferin A at IC50 concentrations for different time intervals and total protein was isolated from the parasites. Briefly, the parasites were resuspended in 180 μl of milliQ water and six freeze thaw cycles were repeated at −80 °C. Further to this lysate, 20 μl of proteinase inhibitor cocktail (1×) was added and sonication at 20% amplitude was performed. The total protein concentration was estimated using the Bradford method. The PTR1 enzyme assay was performed as described above.
Expression analysis of PTR1 transcript in L. donovani parasites
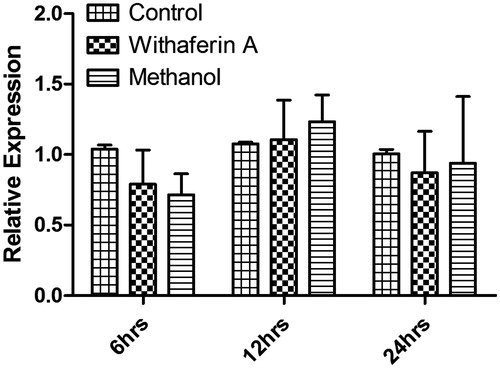
Total RNA was extracted from control and withaferin A treated parasites (5 × 106) at various time intervals using a Nucleospin RNA extraction kit (Machery-Nagel, Deer Park, NY) according to the instructions of the manufacturer. cDNA was synthesized using First Strand cDNA synthesis kit (Takara, Berkeley, CA) from 900 ng of total RNA of each sample. The resulting cDNA was then used for real-time quantitative PCR for PTR1 gene using an ABI 7500 real-time PCR system (Applied Biosystems, Hempstead, UK) with the DNA binding SYBR green dye. β-actin was used as an internal control. Each reaction contained 1 × SYBR Green PCR master mixture (SYBR Green PCR Master Mix; Takara, Berkeley, CA), 10 pmol of each primer and 1.0 μl of cDNA in a final volume of 20 μl. The reaction conditions were as follows: initial activation step (10 min at 94 °C) and cycling step (denaturation for 15 s at 94 °C, annealing for 1 min at 60 °C for 40 cycles) followed by melt curve analysis. The sequence detector software was used for detection of the dequenched probe and calculation of threshold cycles (Ct values) for data analysis. Relative changes in PTR1 mRNA expression were compared with control at respective time points, normalized to β-actin and were quantified by the Δ2−ddCt method. Thus, all the values for experimental samples were represented as relative expression between the sample mRNA and the calibrator (β-actin) mRNA. The data are presented as the means ± SD of data from three independent experiments that yielded similar results.
Results
Effect of withaferin A on L. donovani promastigotes
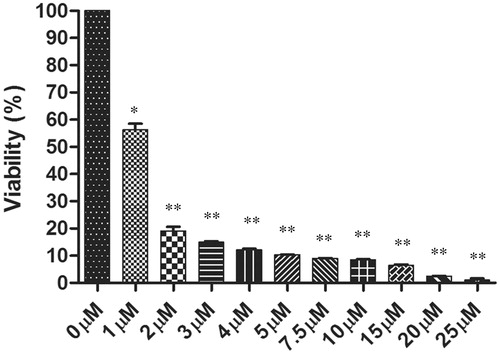
MTT assay was performed to evaluate the antileishmanial effect of withaferin A on the promastigote stage of the parasite. Withaferin A inhibited the growth of parasites in a dose dependent manner and its IC50 concentration was found to be 1.2 μM (). This indicates that withaferin A was able to induce death in the L. donovani promastigotes. The graph is the representative of three independent experiments ± SD.
Figure 1. The antileishmanial effects of withaferin A on promastigotes was determined after 72 h of treatment using MTT assay. The graph depicts the percentage viability of the parasites in the dose dependent concentrations of withaferin A. The values are representative of three values ± SD (*p <0.05, **p <0.01).
In silico docking of withaferin A for PTR1 inhibition
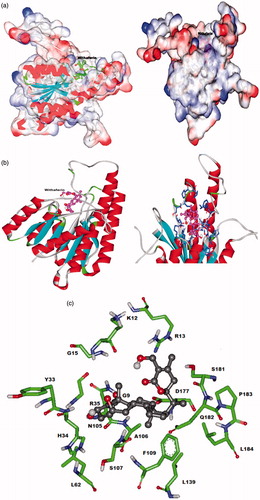
Molecular docking methods are commonly used for predicting the binding modes and energies for proteins or ligandsCitation22. Docking study provided an insight into the prediction of the affinity, activity, binding, and orientation of withaferin A to our target protein molecule LdPTR1. The analysis was based on E-total or free energy of binding and lowest docked energy was calculated. Withaferin A was found to bind LdPTR1 with lowest binding energy of −6.73 kJ/mol. Free binding energy is calculated as the sum of four energy terms namely intermolecular energy (van der Waal, hydrogen bond, desolvation energy, and electrostatic energy), total internal energy, torsional free energy, and unbound system energy. The major interactions shown in the LdPTR1 binding site are the important H-bonds with Gly9, Arg13, His34, and Arg35. The other residues which form a pocket around withaferin A include Gly9, Lys12, Arg13, Gly15, tyr33, his34, Arg35, Leu62, Asn105, Ala106, Ser107, Phe109, Leu139, Asp177, Ser181, Gln182, Pro183, and Leu184. The docking of LdPTR1 and withaferin A in different modes are shown in . Our in silico experiments demonstrate that withaferin A binds to LdPTR1 and may inhibit its function and thus may act as a potential antileishmanial drug.
Figure 2. Predicted interaction model of withaferin A with LdPTR1 (a) space fill model of LdPTR1 with withaferin A docked to the binding site. (b) Red color shows alpha helices (spiral sheets), blue indicates beta sheets, dark blue indicates the residues interacting with withaferin A and pink indicates the withaferin A docked in the binding site. (c) Ball and stick model of the interacting residues in the LdPTR1 with the withaferin A.
Cloning, expression, and purification of LdPTR1
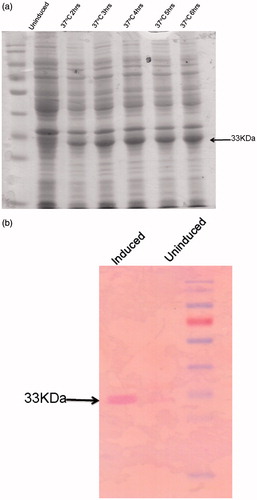
The PTR1 gene was amplified from genomic DNA of DD8 strain of L. donovani and cloned in pET28a vector (). The PTR1 gene encodes for a protein of 288aa with a molecular weight of ≈33 kDa and the plasmid construct was transformed into E. coli BL21 (DE3) bacterial strain. Small-scale cultures of the positive clones (selected on the basis of PCR screening) were subjected to IPTG induction to identify clones capable of expressing the predicted ≈33 kDa recombinant protein. Cultures were induced with 1 mM IPTG at several time points (2–6 h). The protein expression was found to be significantly induced at all the time points considered, the highest being at 4 h. Hence, 4 h induction was considered for further purification studies (). Expression of recombinant protein was also validated by western blot analysis using anti-His antibody (Sigma, St. Louis, MO). The predicted ≈33 kDa recombinant protein band was visualized on the membrane using BCPIP/NBT reagent () that also indicates the recombinant protein was in frame with His-tag.
Figure 3. Agarose gel electrophoresis of the LdPTR1 gene product. (a) The figure shows the 867 bps amplified LdPTR1 product from the genomic DNA along with DNA ladder. (b) Restriction digestion of the positive clone plasmid showing the free insert (867 bps) and the plasmid (5.5 kb) with DNA ladder.
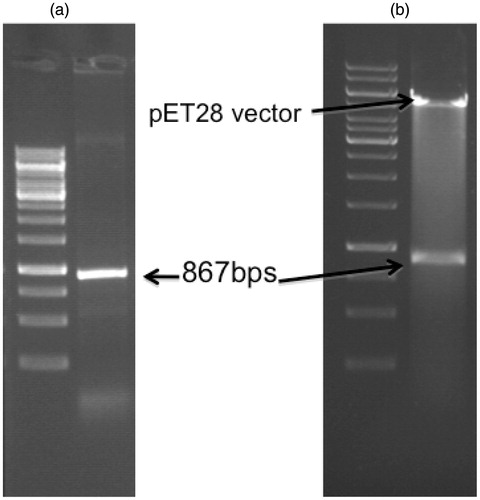
Figure 4. Induction and western blot of LdPTR1. (a) Proteins were separated on 12% SDS-PAGE before and after induction with 1 mM IPTG at different time points. The induced LdPTR1 (33 kDa) was represented in the figure. (b) Western blot analysis using anti-his antibody using the uninduced and induced crude lysate along with protein marker. The blot was developed using anti mouse ALP conjugate.
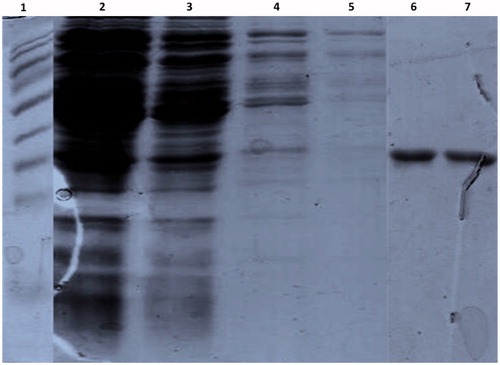
The relative distribution of protein of interest was examined in both soluble and insoluble fractions of bacterial cell lysates. After the sonication, both the fractions obtained were subjected to SDS-PAGE to detect the recombinant protein and almost 80% of recombinant protein was observed to present in the supernatant fraction (data not shown). The recombinant protein was purified based on its His6 tag by affinity chromatography using a Ni2+ chelating Sepharose column as described in Materials and methods section. The purification conditions were standardized using increasing concentrations of imidazole (50–200 mM) in the elution buffer after three washes with wash buffer containing 20 mM imidazole. The pre-equilibration step of Ni2+ column with wash buffer containing 20 mM Imidazole removed all the non-specifically bound proteins. In addition, elutions by imidazole gradient facilitated the release of more tightly bound non-specific proteins in the early fractions of elution. The purity of the desired protein was more than 95% in 200 mM imidazole elution and further large-scale purification was done using the same imidazole concentration (). The various eluted fractions at 200 mM imidazole concentration were pooled and dialyzed overnight at 4 °C in 1 L of dialysis buffer that had been changed for every 6 h intervals. The purified protein was stored at −80 °C for further studies.
Figure 5. The expressed His-tag fusion protein was purified using a Ni2+ chelating Sepharose column affinity chromatography. The purified proteins were separated on 12% SDS-PAGE and stained with coomassie blue. Lane 1: protein marker, Lane 2: flow through, Lane 3–5: washes with the wash buffer, Lane 6 and 7: Elution of the protein with 200 mM imidazole.
Enzyme inhibition studies of recombinant PTR1 with withaferin A
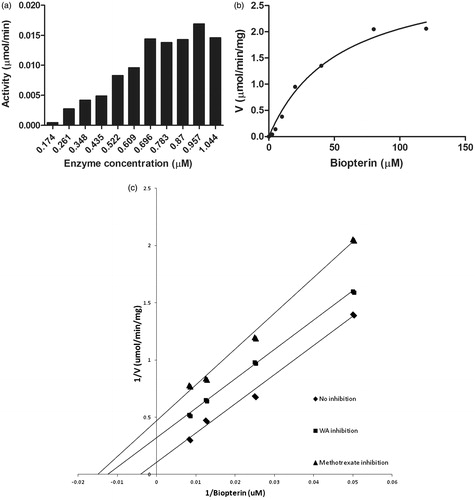
PTR1 enzyme kinetics was performed using the Michaelis–Menten reaction. First, we optimized the enzyme concentration for carrying out the reaction using biopterin (100 μM) as a substrate and NADPH (100 μM) as a co-factor to determine the Vmax and Km for the biopterin substrate. The velocity of PTR1 increased up to 0.696 μM of the substrate after which there was no change in the velocity of the reaction with increase in the substrate concentration (). Hence, we used 0.696 μM as the optimum enzyme concentration for further reactions. The enzyme assay was performed with various concentrations of substrate, 6-biopterin (0–120 μM) and found the Vmax and Km of the reaction to be 2.5 μmol/min/mg and 35 μM, respectively (). The enzyme kinetics was performed using withaferin A as an inhibitor and using the Lineweaver–Burk plot, the Ki value was determined to be 0.9 μM and using methotrexate (a positive control) as an inhibitor, the Ki value was found to be 1.126 μM. (). It followed an uncompetitive mode of inhibition.
Figure 6. Enzyme kinetics of purified LdPTR1. (a) The optimum concentration of the enzyme was carried using 100 μM NADPH and 100 μM biopterin. It was found to be 0.696 μM, which was used for further assays. (b) The recombinant LdPTR1 enzyme assay was performed using 20 mM sodium acetate buffer (pH 4.8) for the different concentrations of substrate biopterin. (c) The enzyme inhibition studies were performed for methotrexate and withaferin A using lineweaver Burk plot. The Ki values for each compound were calculated from the graph. The experiments were repeated three times and figure represents the result from one such experiment.
Enzyme inhibition assay on parasite crude lysates
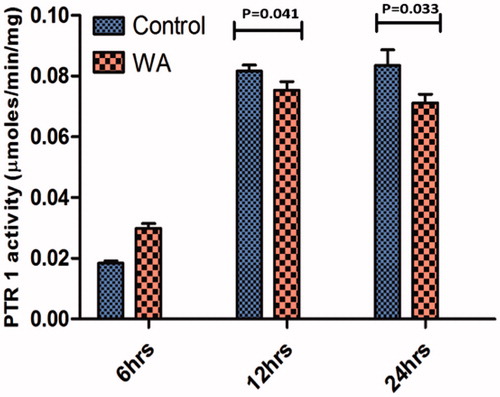
To prove the PTR1 enzyme inhibition in the parasites upon withaferin A treatment, we performed the PTR1 enzyme assay in the total protein lysates isolated from untreated and withaferin A-treated parasites. Although there was no significant difference at 6 h treatment, the enzyme inhibition was observed to increase with time. At 12 h, the inhibition was more significant than 6 h treatment (p = 0.041) whereas at 24 h, the enzyme inhibition was much more significant (p = 0.033). The experiment was repeated three times and data represents the mean of three independent experimental values ± SD (). This reveals that withaferin A indeed inhibits the LdPTR1 enzyme in vitro.
Figure 7. PTR1 enzyme activity was determined in the crude parasite lysates of control and withaferin A treated at different time intervals. The values are representative of three independent experiments and significant values indicate the comparison of control with treated samples at that particular time point.
Endogenous mRNA expression levels of PTR1
To assess the expression of LdPTR1 in the control and treated parasites, total RNA was isolated and cDNA was prepared. The mRNA expression levels were determined by quantitative real-time PCR analysis and were found to be unaltered between control and withaferin A treated parasites at all time points. The expression levels were also unchanged in the methanol treated parasites compared with control. The above results show that withaferin A is not altering the endogenous expression levels of PTR1 instead it might be showing direct binding effect on the PTR1 protein ().
Discussion
During the twenty-first century, the plant-based compounds have gathered much importance in the trials of discovering new antileishmanial drugs with high efficacy and fewer side effects. Our former study has shown that withanolides extracted from W. somnifera leaves exhibit potent antileishmanial activity against the promastigote stage of L. donovani parasites, but the mode of action of these withanolides has not been elucidated yetCitation6. A previous study in the L. donovani showed that withaferin-A inhibits the protein kinase, which induces apoptosis through apoptotic topoisomerase I-DNA complexCitation23. A recent study also showed that the antileishmanial activity of the herbal drug from W. somnifera is exhibited through blocking of protein kinase C signaling pathwayCitation24. Till now, no report has demonstrated its antileishmanial activity on any of the important pathways that are essential for the growth of the parasite. Our current study focuses on the mode of action of withaferin A, an abundant withanolide in the leaves of W. somnifera, on the Pteridine salvage enzyme of the parasite pterdine reductase 1.
The novel pathways exclusively present in trypanosomatids would provide for excellent drug targets among which pteridine pathway stands one. Leishmania donovani parasites are auxotrophs for pteridines that play a major role in reducing pteridines and also functions as a metabolic bypass for DHFR-TS by reducing the folates. Thus, an inhibitor targeting pteridine reductase 1 could also possibly target the DHFR-TS enzyme due to the structural similarity of the substrates. A single compound might be found with good efficacy against both enzymes due to the structural similarities of the substratesCitation7,Citation8,Citation25,Citation26. The reduced bioavailability of pterins makes the parasites more susceptible to the oxidant damage, which correlates with our previous study that showed the enhancement of the production of cellular reactive oxygen species.
In this regard, we determined the effect of withaferin A on promastigote stage of the parasite and it was found to exhibit the antileishmanial effect in a dose-dependent manner. The IC50 value was determined to be 1.2 μM from the graph. To decipher its mode of action on the parasites, we performed an in silico docking analysis of parasite’s pteridine reductase 1 enzyme with withaferin A, which revealed its binding residues. Previous docking study with monastrol by Kaur et al.Citation27 revealed that hydrophobic residues namely Arg17, Asn109, Ser111, Asp181, Tyr191, Tyr194, Lys198, Leu226, and Ala230 mainly enfold the active sites of LdPTR1, but withaferin A binds with LdPTR1 and forms hydrogen bonding with Gly9, Arg13, His34, and Arg35 whereas the residues involved in forming a pocket around the withaferin A are Gly9, Lys12, Arg13, Gly15, Tyr33, His34, Arg35, Leu62, Asn105, Ala106, Ser107, Phe109, Leu139, Asp177, Ser181, Gln182, Pro183, and Leu184. Surprisingly, we did not find any cysteine residues in LdPTR1 interacting with withaferin A as hydrophobic interactions of withaferin A involve this residueCitation28. The binding energy of withaferin A with LdPTR1 was found to be −6.73 kJ/mol. The determined high binding affinity of withaferin A with LdPTR1 might provide an insinuation for the mechanism of biological activity of withaferin A.
Subsequently, the PTR1 gene was amplified from DD8 strain of L. donovani and cloned into pET28a vector. The protein was expressed in E. coli BL21 (DE3). The protein was purified from the soluble fraction through Ni2 + chelating Sepharose columnCitation29. In the gradient elution of 50–200 mM imidazole, the peak fraction was eluted at 200 mM imidazole concentration containing with the purity of protein more than 95%.
The docking results were validated using the inhibition studies of recombinant enzyme LdPTR1 with withaferin A. The optimal concentration of LdPTR1 for its activity was found to be 0.696 μM. We have used the sodium acetate buffer pH 4.8 for determining the activity of LdPTR1 as previously reportedCitation30. The subsequent assays were performed at this optimum pH and enzyme concentration. We estimated the PTR1 activity with oxidized biopterin using standard the Michaelis–Menten kinetics. Km and Vmax values for biopterin substrate were derived from the Lineweaver–Burk plot. Methotrexate is a known antifolate inhibitor of Plasmodium falciparum DHFRCitation31. We used methotrexate as an inhibitor for LdPTR1 and found its Ki value as 1.126 μM against the substrate biopterin. Further inhibition studies of LdPTR1 performed with withaferin A yielded a Ki value of 0.9 μM. In addition to this, the time-dependent inhibition (at IC50 concentration) of LdPTR1 in withfarein A-treated parasite lysates suggests its inhibitory role in promastigote stage in vitro, but withaferin A did not have any inhibitory role in the endogenous expression of PTR1. This might be due to the independent role of pterins in Leishmania growth apart from folate biosynthesisCitation7,Citation8. Additional preclinical experiments of withaferin A in mice model for visceral infection would shed light on the mode of action in vivo.
Conclusions
Withaferin A has been reported for its anticancer properties in various cancer types. Herein, we demonstrated that the antileishmanial activity of withaferin A might be through the inhibition of Pteridine reductase 1 enzyme of L. donovani. Further studies advocate the usage of withaferin A in treating this dreadful disease either individually or in combination with current antileishmanial drugs.
Declaration of interest
The authors report that they have no conflicts of interests. Indian Council of Medical Research (ICMR), New Delhi, India, has supported the work and Department of Animal Biology and School of Life Sciences, University of Hyderabad provided equipment facilities. Financial assistance in the form of fellowship from Council of Scientific and Industrial Research (CSIR) is greatly acknowledged. R. K. G. wants to thank D.S. Kothari Post Doctoral Fellowship, University Grants Commission, Government of India (No. F. 4-2/2006 (BSR)/13–618/2012(BSR) is for the grant of Post Doctoral Fellowship.
References
- Desjeux P. Human leishmaniases: epidemiology and public health aspects. World Health Stat Q 1999;45:267–75
- Desjeux P. Leishmaniasis: current situation and new perspectives. Comp Immunol Microbiol Infect Dis 2004;27:305–18
- Sundar S, More DK, Singh MK, et al Failure of pentavalent antimony in visceral leishmaniasis in India: report from the Center of the Indian Epidemic. Clin Infect Dis 2000;31:1104–7
- Thakur CP, Thakur S, Narayan S, Sinha A. Comparison of treatment regimens of kala-azar based on culture & sensitivity of amastigotes to sodium antimony gluconate. Indian J Med Res 2008;127:582–8
- Sundar S, Singh A, Rai M, et al Efficacy of miltefosine in the treatment of visceral leishmaniasis in India after a decade of use. Clin Infect Dis 2012;55:543–50
- Chandrasekaran S, Dayakar A, Veronica J, et al An in vitro study of apoptotic like death in Leishmania donovani promastigotes by withanolides Leishmania promastigotes withanolides. Parasitol Int 2013;62:253–61
- Nare B, Luba J, Hardy LW, Beverley SM. New approaches to Leishmania chemotherapy: pteridine reductase 1 (PTR1) as a target and modulator of antifolate sensitivity. Parasitology 1997;114:S101–10
- Nare B, Hardy L, Beverley SM. The roles of pteridine reductase 1 and dihydrofolate reductase-thymidylate synthase in pteridine metabolism in the protozoan parasite Leishmania major. J Biol Chem 1997;272:13883–91
- Bello AR, Nare B, Freedman D, et al PTR1: a reductase mediating salvage of oxidized pteridines and methotrexate resistance in the protozoan parasite Leishmania major. Proc Natl Acad Sci USA 1994;91:11442–6
- Sienkiewicz N, Ong HB, Fairlamb AH. Trypanosoma brucei pteridine reductase 1 is essential for survival in vitro and for virulence in mice. Mol Microbiol 2010;77:658–71
- Wang J, Leblanc E, Chang CF, et al Pterin and folate reduction by the Leishmania tarentolae H locus short-chain dehydrogenase/reductase PTR1. Arch Biochem Biophys 1997;342:197–202
- Hardy LW, Matthews W, Nare B, Beverley SM. Biochemical and genetic tests for inhibitors of Leishmania pteridine pathways. Exp Parasitol 1997;87:157–69
- Weiner WA, Weiner J. Ashwagandha (Indian ginseng). In: Herbs that heal. Mill Valley (CA): Quantum Books; 1994:70–2
- Matsuda H, Murakami T, Kishi A, Yoshikawa M. Structures of withanosides I, II, III, IV, V, VI, and VII, new withanolide glycosides, from the roots of Indian Withania somnifera DUNAL and inhibitory activity for tachyphylaxis to clonidine in isolated guinea-pig ileum. Bioorg Med Chem 2001;9:1499–507
- Mathur R, Gupta SK, Singh N, et al Evaluation of the effect of Withania somnifera root extracts on cell cycle and angiogenesis. J Ethnopharmacol 2006;105:336–41
- Shah N, Kataria H, Kaul SC, et al Effect of the alcoholic extract of Ashwagandha leaves and its components on proliferation, migration, and differentiation of glioblastoma cells: combinational approach for enhanced differentiation. Cancer Sci 2009;100:1740–7
- Malik F, Kumar A, Bhushan S, et al Immune modulation and apoptosis induction: two sides of antitumoural activity of a standardised herbal formulation of Withania somnifera. Eur J Cancer 2009;45:1494–509
- Widodo N, Priyandoko D, Shah N, et al Selective killing of cancer cells by Ashwagandha leaf extract and its component withanone involves ROS signaling. PLoS One 2010;5:e13536
- Malik F, Kumar A, Bhushan S, et al Reactive oxygen species generation and mitochondrial dysfunction in the apoptotic cell death of human myeloid leukemia HL-60 cells by a dietary compound withaferin A with concomitant protection by N-acetyl cysteine. Apoptosis 2007;12:2115–33
- Stan SD, Zeng Y, Singh SV. Ayurvedic medicine constituent withaferin A causes G2 and M phase cell cycle arrest in human breast cancer cells. Nutr Cancer 2008;1:51–60
- Oh JH, Lee TJ, Kim SH, et al Induction of apoptosis by withaferin A in human leukemia U937 cells through down-regulation of Akt phosphorylation. Apoptosis 2008;13:1494–504
- Bikadi H, Hazai E. Application of the PM6 semi-empirical method to modeling proteins enhances docking accuracy of AutoDock. J Cheminform 2009;11:1–15
- Sen N, Banerjee B, Das BB, et al Apoptosis is induced in leishmanial cells by a novel protein kinase inhibitor withaferin A and is facilitated by apoptotic topoisomerase I-DNA complex. Cell Death Differ 2007;14:358–67
- Grover A, Katiyar SP, Jeyakanthan J, et al Blocking protein kinase C signaling pathway: mechanistic insights into the anti-leishmanial activity of prospective herbal drugs from Withania somnifera. BMC Genomics 2012;13:S20
- Knighton DR, Kan CC, Howland E, et al Structure of and kinetic channelling in bifunctional dihydrofolate reductase-thymidylate synthase. Nat Struct Biol 1998;1:186–94
- Zuccotto F, Martin AC, Laskowski RA, et al Dihydrofolate reductase: a potential drug target in trypanosomes and leishmania. J Comput Aided Mol Des 1998;12:241–57
- Kaur J, Sundar S, Singh N. Molecular docking, structure–activity relationship and biological evaluation of the anticancer drug monastrol as a pteridine reductase inhibitor in a clinical isolate of Leishmania donovani. J Antimicrob Chemother 2010;65:1742–8
- Antony ML, Lee J, Hahm ER, et al Growth arrest by the antitumor steroidal lactone withaferin A in human breast cancer cells is associated with down regulation and covalent binding at cysteine 303 of β-tubulin. J Biol Chem 2014;289:1852–65
- Kumar P, Kothari H, Singh N. Overexpression in Escherichia coli and purification of pteridine reductase (PTR1) from a clinical isolate of Leishmania donovani. Protein Expr Purif 2004;38:228–36
- Kaur J, Kumar P, Tyagi S, et al In silico screening, structure–activity relationship, and biologic evaluation of selective pteridine reductase inhibitors targeting visceral leishmaniasis. Antimicrob Agents Chemother 2011;55:659–66
- Shallom S, Zhang K, Jiang L, Rathod PK. Essential protein-protein interactions between Plasmodium falciparum thymidylate synthase and dihydrofolate reductase domains. J Biol Chem 1999;274:37781–6