Abstract
Due to a poor clinical predictive power of 2D cell cultures, standard tool for in vitro assays in drug discovery process, there is increasing interest in developing 3D in vitro cell cultures, biologically relevant assay feasible for the development of robust preclinical anti-cancer drug screening platforms. Herein, we tested amidino-substituted benzimidazoles and benzimidazo[1,2-a]quinolines as a small platform for comparison of antitumor activity in 2D and 3D cell culture systems and correlation with structure–activity relationship. 3D cell culture method was applied on a human cancer breast (SK-BR-3, MDA-MB-231, T-47D) and pancreatic cancer cells (MIA PaCa-2, PANC-1). Results obtained in 2D and 3D models were highly comparable, but in some cases we have observed significant disagreement indicating that some prominent compounds can be discarded in early phase of researching because of compounds with false positive result. To confirm which of cell culture systems is more accurate, in vivo profiling is needed.
Introduction
Analyses of the clinical trials reveal that ∼67% of drug leads molecules fail in late clinical trials stages. Major causes of human trials failure are insufficient efficacy and safety issuesCitation1. The largest number of failures is in the oncology field.
One of the main reasons of high drug attrition rate in oncology is poor clinical predictive power of 2D cell cultures that biologist use as standard tool for in vitro assaysCitation2. Conventional 2D cell cultures are not able to completely describe a malignant phenotype related to tumorogenicity in vivo. In 2D culture conditions, cells need to adjust to a flat and rigid surface that can result in altered cell metabolism and functionality. Thus, cellular functions and physiological responses that are present in organs are often lost in 2D cell cultures which are usual standard for testing new chemical entities (NCE). That influence ability of an in vitro assay to produce reliable information about tested NCE’s which is essential in early phase of drug development. Contrary, 3D cell culture techniques have become increasingly popular and are suggested to be better models than 2D monolayers due to improved cell-to-cell contact and structures that resemble in vivo architecture. Cell shape and contacts between cells influence the cytoskeleton which can regulate gene and protein expression and hence cell functionCitation4. Once expensive, laborious and difficult to be used for screening of numerous novel chemical entities, 3D spheroid cultures technique is today applicable for both basic research and high-throughput screening (HTS).
Nevertheless, the ideal method for 3D cell culture is not yet optimised. Among various 3D methods, hanging drop method is one of the most well-characterised models for 3D culture with all the advantages and disadvantagesCitation5. In our research, we have used 3D Biomatrix hanging drop plates (http://www.3dbiomatrx.com/) because they support growing of 3D cell culture on a larger scale and also HTS of NCECitation6.
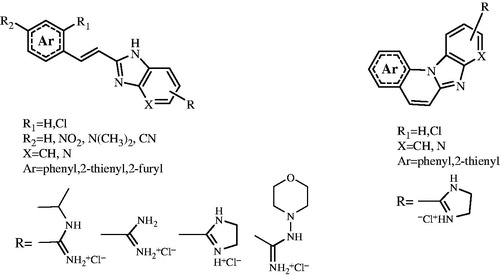
We screened 22 compounds () that were designed and prepared as prominent examples of continuous scientific research in the field of potential, biologically active benzimidazoles and used them as a small platform to compare potential antitumor activity in 2D and 3D cell culture systems. The constant and growing interest over the past few years for the synthesis and biological studies of substituted benzimidazoles and their azino-fused benzannulated derivatives is owed partly to their well-known biological activities such as anticancer, antimicrobial, antihistaminic, antiviral, antianalgestic, antifungal and many others.
Synthesis and biological activity of several groups of benzimidazo[1,2-a]quinolines, including positively charged amidino-substituted benzimidazo[1,2-a]quinolines and their heteroaromatic analogues was previously reportedCitation7–9. Also biological studies that confirmed the anticancer potential of this class of compounds, especially that of positively charged amidino-substituted analogues of benzimidazo[1,2-a]quinolines which may intercalate into double-stranded DNA or RNACitation10.
Our goal was to compare the drug sensitivities of the three human breast cancer cell lines (SK-BR-3, MDA-MB-231, T-47D) and two pancreatic cancer cell lines (MIA PaCa-2 and PANC-1) grown in 2D and 3D cell culture conditions and test the hypothesis about differences in the activities of chemotherapeutic drugs in 2D and 3D assaysCitation11.
Chemistry
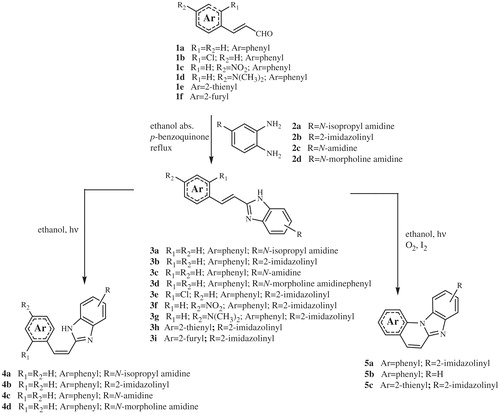
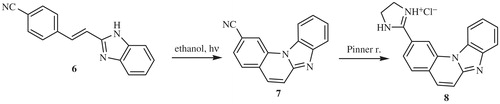
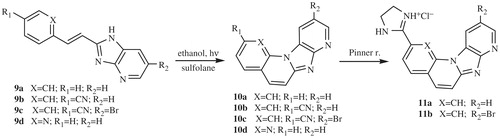
The majority of compounds related to benzimidazoles and benzimidazo[1,2-a]quinolines were previously synthesised and spectroscopically characterised by our research groupCitation7–9. Amidino substituted E-acyclic benzimidazole derivatives 3a–3i were prepared in the condensation of aromatic propenales 1a–1d, 1e and 1f with 4-N-amidino-substituted 1,2-phenylenediamines 2a–2d. Z-acyclic benzimidazole derivatives 4a–4d were prepared by photochemical isomerisation from corresponding E-isomers while cyclic benzimidazo[1,2-a]quinoline derivatives 5a–5b and its 2-thienyl analogue 5c were prepared either by photochemical dehydrocyclization or termic dehydrohalogenation reaction in sulfolane at high temperature (Scheme 1). 2-(2-Imidazolinyl) benzimidazo[1,2-a]quinoline 8 was prepared from cyano-substituted precursor 7 in the Pinner reaction according to the Scheme 2. Triaza-benzo[c]fluorenes 10a–10d were prepared by photochemical or termic dehydrocyclization from acyclic imidazo[4,5-b]pyridine derivatives 9a–9d. 2-Imidazolinyl-substituted triaza-benzo[c]fluorenes 11a–11b were prepared using the Pinner reaction from the corresponding cyano-substituted precursors (Scheme 3).
Setup for high-throughput 2D and 3D drug screening
To compare the difference in responses in 3D and 2D cell culture systems we have chosen four cytostatic drugs (vinorelbine, docetaxel, doxorubicine and gemcitabine) with different mode of action as a standard compounds. These will provide a more reliable comparison of culture model-influenced drug sensitivities. Compounds were screened in traditional 2D model and 3D Biomatrix model on three human breast cancer cell lines (SK-BR-3, MDA-MB-231 and T-47D) and two pancreatic cancer cell lines (MIA PaCa-2 and PANC-1). Amidino substituted benzimidazole and benzimidazo[1,2-a]quinoline derivatives (22 prepared compounds) were tested in selected breast and pancreatic cancer cells in 2D cell culture system (). After screening of cells in 2D assay we have selected a mixture of 13 active and inactive compounds based on variations in structure for additional testing in 3D cell culture. Antitumor activity was measured by MTS viability assayCitation12. For 2D cell culture assay, cells were grown in 96 well cell star polystyrene plates, 10 000/cell/well, while for 3D cell culture assay, cells were grown in 96 well Perfecta 3D hanging drop plates 5000/cell/well. In order to determine the influence of testing chemotherapeutic substances on the cell viability in both formats, assay were firstly performed by culturing three cell lines in 2D format for 24, 48 and 72 h. Cells were seeded in wells 4 h prior the treatment with compounds. Following results, the 72-h time point was chosen to be a culturing time for 3D format to ensure the required time for a spheroid growth of the cells. In both types of the assay, cells were seeded per well, 4 h prior the treatment with different substances concentrations. Therefore, cells were seeded in wells and after 4 days formation of spheres were checked under microscope. After this, compounds were added in different concentrations and incubated for 72 h. A cell viability assay was performed according to the manufacturer’s instruction, using cell titre 96 aqueous solution, MTS kit (Promega, Madison, WI). After 0.5–2 h of cell incubation with MTS, the plates were read using PE EnVision at 490 nm. The results for each of tested compounds are reported as growth percentages from two independent concentrations curves compared with the untreated control cells after drug exposure.
Table 1. IC50 of 22 compounds on 2D assay.
Antitumor activity for each compound is presented as IC50 value. IC50 values were calculated using the programme GraFit 5.0.12 (Surrey, UK), and average values from three independent experiments.
Experiment section
Chemistry
General methods
Melting points were determinated on a Koffler hot stage microscope and are uncorrected. IR spectra were recorded on a Perkin-Elmer Spectrum 1 spectrophotometer, with KBr disks. 1H and 13C NMR spectra were recorded on Bruker Avance DPX 300 and Bruker Avance DRX 500 spectrometers using TMS as an internal standard in DMSO-d6. Mass spectra were recorded on Agilent 1100 series LC/MSD Trap SL. Elemental analysis for carbon, hydrogen and nitrogen were performed on a Perkin-Elmer 2400 elemental analyser and a Perkin-Elmer, Series II, CHNS Analyser 2400 (Shelton, CT). Where analyses are indicated only as symbols of elements, analytical results obtained are within 0.4% of the theoretical value. All compounds were routinely checked by TLC with Merck silica gel 60F-254 glass plates (Darmstadt, Germany).
Synthesis
The synthesis and spectroscopic characterisation of compounds 3a–3e, 3 –3i, 4a–4d, 5a–5c, 6–8, 9a–9d, 10a–10d and 11a–11b was previously published in our research group.Citation7–9
General method for the synthesis of 5(6)-N-(2-imidazolinyl)-substituted E-2-styryl-1H-benzimidazoles (3f, 3g)
A mixture of 4-(2-imidazolinyl)-1,2-phenylenediamine hydrochloride, corresponding 3-phenyl-propenal and p-benzoquinone in absolute ethanol was stirred at reflux for 3 h under nitrogen atmosphere. The reaction mixture was cooled to room temperature and diethyl ether was added. Resulting product was filtered off and washed with diethylether. After recrystallisation from ethanol/diethyl ether or ethanol/acetone, powders were obtained.
E-(2-imidazolinyl)-2–(4-nitrostyryl)benzimidazole-5(6)-carboxamidine hydrochloride (3f)
Compound 3f was prepared using the general method described for the preparation of 3f and 3g; a mixture of 4-(2-imidazolinyl)-1,2-phenylenediamine hydrochloride (0.240 g, 1.13 mmol), 3–(4-nitrophenyl)propenal (0.200 g, 1.13 mmol) and p-benzoquinone (0.122 g, 1.13 mmol) in absolute ethanol (10 ml) was refluxed under nitrogen for 2 h and then reaction mixture was worked up to give 0.260 g (60%) of yellow powder; mp >290 °C; IR (KBr) (νmax/cm−1) 3430, 2912, 1700, 1611; 1H NMR (DMSO-d6) (δ/ppm) 13.58 (bs, 1H, NHbenzimidazole), 10.40 (bs, 2H, NHamidine), 8.35 (s, 1H, Harom.), 8.30 (d, 2H, J = 8.82 Hz, Harom.), 7.98 (d. 1H, J = 16.80 Hz, Hethenyl.), 7.93 (d, 2H, J = 8.76 Hz, Harom.), 7.84 (d, 1H, J = 7.66 Hz, Harom.), 7.79 (d, 1H, J = 7.56 Hz, Harom.), 7.52 (d, 1H, J = 16.88 Hz, Hethenyl), 4.04 (s, 4H, CH2); 13C NMR (DMSO-d6) (δ/ppm): 163.0, 149.8, 143.2, 141.8, 136.6, 129.0, 127.1, 123.5, 122.2, 122.0 (s), 120.7, 117.6, 115.4 (2C), 111.3, 44.90, 21.4; MS (m/z): [M + H]+ = 334; Anal. (C18H16N5O2Cl) Calc. C 58.46, H 4.36, N 18.94%; Found C 58.77, H 4.42, N 19.20%.
E-(2-imidazolinyl)-2–(4-N,N-dimethylamino)styryl-3H-benzimidazole-5(6)-carboxamidine hydrochloride (3g)
Compound 3g was prepared using the general method described for the preparation of 3f; a mixture of 4-(2-imidazolinyl)-1,2-phenylenediamine hydrochloride (0.352 g, 1.66 mmol), 3–(4-N,N-dimethylaminophenyl)propenal (0.250 g, 1.66 mmol) and p-benzoquinone (0.179 g, 1.66 mmol) in absolute ethanol (10 ml) was refluxed under nitrogen for 2 h and then reaction mixture was worked up as it is described to give 0.240 g (39%) of yellow powder; mp >290 °C; IR (KBr) (νmax/cm−1) 3434, 2911, 1690, 1616; 1H NMR (DMSO-d6) (δ/ppm) 13.20 (bs, 1H, NHbenzimidazole), 10.55 (bs, 2H, NHamidine), 8.26 (s, 1H, Harom.), 7.77 (d, 1H, J = 16.60 Hz, Hethenyl), 7.75 (d, 2H, J = 8.50 Hz, Harom.), 7.72 (d, 1H, J = 7.92 Hz, Harom.), 7.50 (d, 2H, J = 8.65 Hz, Harom.), 6.95 (d, 1H, J = 16.58 Hz, Hethenyl.), 6.75 (d, 2H, J = 8.65 Hz, Harom), 4.05 (s, 4H, CH2), 3.23 (s, 6H, CH3); 13C NMR (DMSO-d6) (δ/ppm): 164.1, 150.0, 143.8, 142.0, 138.1, 129.5, 127.5, 123.7, 122.4, 122.4 (s), 121.2, 118.3, 116.1 (2C), 111.5, 45.9, 43.3 (2C), 23.4; MS (m/z): [M + H]+ = 332; Anal. (C20H22N5Cl) Calc. C 65.30, H 6.03, N 19.04%; Found C 64.88, H 6.27, N 19.22%.
Antitumor activity assays
Material and methods
Test compounds
Doxorubicin was purchased from Apollo (BID 0120; Opelika, AL) , gemcitabine, docetaxel and vinorelbine were purchased from TOCRIS (Cat 3259, 4056 and 3401; Bristol, UK). Test compounds were synthesised by research group at Department of Organic Chemistry, Faculty of Chemical Engineering and Technology, University of Zagreb.
Mother plates (384-deep-well-V plates Perkin-Elmer, Cat. 6008590) with serial dilutions of compounds in pure DMSO are prepared from 6 mM DMSO stock solutions on Janus automatic pipetting workstation (Perkin-Elmer). Compounds are diluted 1:3. 500 nl of compound were transferred from mother plate to test plate by using Mosquito (TTP labtech). DMSO percentage in test concentrations was <0.3%. Start concentration of the standard compounds and test compounds was 30 μM.
Cell cultures
Human breast cancer cell lines SK-BR-3 (HTB-30), MDA-MB-231 (HTB-26) and T-47D (HTB-133) and human prostate cancer cell lines MIA PaCa-2 and PANC-1 were purchased from ATTC and were maintained in appropriated, recommended by supplier medium, supplemented with 10% heat inactivated foetal bovine serum and penicillin/streptomycin/amphotericin B, in a humidified atmosphere of 5% CO2 and 95% O2 at 37 °C.
Statistical analysis
Calculation of IC50 data, curves and QC analysis is made by using Excel tools and GraphPadPrism software (La Jolla, CA), v. 5.03. In brief, individual concentration–effect curves are generated by plotting the logarithm of the tested concentration of tested compounds (X) versus corresponding percent inhibition values (Y) using least squares (ordinary) fit. Best fit IC50 values are calculated using Log(inhibitor) versus normalised response − Variable slope equation, where Y = 100/(1 + 10((LogIC50 − X) * HillSlope)). QC criteria parameters (Z′, S:B, R2, HillSlope) were checked for every IC50 curve.
Results and discussion
Differences in drug responses between 2D and 3D culture models
We reported the variations in responses of the standard drugs () and tested compounds in 2D and 3D cell culture systems ( and ).
Table 2. IC50 of standard drugs on 2D and 3D assay.
Table 3. Comparison of IC50 of selected NCE on 3D and 2D assay.
As shown in , almost for all tested standard drugs in 2D assay, IC50 values decreased depending on the incubation time. The standard drugs achieved the highest activity after 72 h of incubation in comparison with 24 and 48 h of incubation. Therefore, we compare IC50 values obtained in the 2D and 3D assay at this time point. Exceptions were PANC-1 and SK-BR-3 cell lines that showed high gemcitabine resistance. The obtained result was comparable with IC50 values found in the literatureCitation13,Citation14 for standard drugs in 2D assay, taking in account the differences in assay setup (number of cells per well, source and formulation of tested standard drugs, washout period, etc.)Citation15.
Comparison of IC50 values from 3D assay and IC50 values from 2D assay after 72 h of incubation showed a shift in IC50 values: all cell lines showed increased chemoresistence for vinorelbine and docetaxel in 3D in comparison to 2D cell culture system (3D IC50 > 2D IC50); in 3D assays doxorubicine also showed higher IC50 values on PANC-1 and SK-BR-3 cell lines and lower or equal values on MIA-PA-Ca-2, MDA-MB-231 and T47D (3D IC50 ≤ 2D IC50); gemcitabine showed in the 3D assay higher IC50 value for MIA-PA-Ca-2 and a lower IC50 value for MDA-MB-231 and T47D. We concluded that, as well as in 2D assay, the shift in IC50 in a 3D cell culture models is not dependent only on the mode of action of the tested compounds, but it is also affected by the cell line sensitivity and in case of 3D cultures spheroid growth has an impact on the cell line sensitivity. A correlation between IC50 value and mechanism of action of the standard drugs was not obtained.
Structure–activity relationship (SAR) in 2D assays
SAR in 2D assays () reveals that activity of tested compounds is strongly depended on rigidity of central core and the nature of the functional group on benzimidazole ring or phenyl ring. The first set of tested compounds, acyclic amidino substituted E-2-styrylbenzimidazole derivatives 3a–3i, proved to be mainly inactive or moderate active on all tested cell lines in the 2D assays. Moderate activity was observed only with compounds 3f and 3 g, which contains polar functionality in structure. In fact, compound 3g shows moderate activity even in the pancreatic cell line MIA-Pa-Ca-2. The acyclic Z-2-styrylbenzimidazole analogues 4a–4d were inactive on all 2D assays.
Obviously the non-planar conformation prevents binding of compounds with target. However, significant improvement on activity was obtained by cyclisation and formation of rigid system in compounds 5a and 5c, but not in 5b.
Once again, the prerequisite of 2-imidazolinyl group was confirmed by the significant difference between inactive compound 5b and 2-imidazolinyl active compounds 5a and 5c. Substitution of phenyl ring in 5a with 2-thienyl group in 5c did not affect the activity. Guided by the positive results we have extended the SAR with the new structures that contains a 2-imidazolinyl group. Translation of the 2-imidazolinyl group found on benzimidazole ring in 5a to a phenyl ring in 8 did not change activity significantly. Further introduction of an additional nitrogen atom in structure of 8 gave compound 11a, which showed an increase in activity on all cell lines, especially on T47D. This positive result was also beneficial for decreasing the compounds lipophilicity. Finally, the bromo-substituted tetracycle 11b showed the most pronounced activity towards PANC-1 and MDA-MB-231 cell lines into the submicromolar range of potency. In line with the previous results, the compounds 10a–10d as analogues of 11a–11b without 2-imidazolinyl group were either inactive or poorly active on all tested cell lines.
SAR in 3D assays
The results of antitumor activity of compounds in the 3D cell culture system are, in most cases, highly comparable with results obtained in the 2D cell culture system for acyclic E-2-styrylbenzimidazoles 3a–3f (). The exception is compound 3g that was moderately active on MIA-Pa-Ca-2 and T47D cells in the 2D assays, was inactive on MIA-Pa-Ca-2 cells and highly active in T47D cells in the 3D assays. Disagreement in activity was also observed with compounds 5a and 8 that differ only in position of 2-imidazolinyl group. Named 2-imidazolinyl group found on benzimidazole ring in 5a was translated to a phenyl ring in 8. Both compounds 5a and 8 showed moderate and comparable activities in the 2D cell assays on all tested cell lines. However, in 3D assay compound 5a showed pronounced activity towards PANC-1 and no activity on SK-BR-3 cell line, while compound 8 proved to be inactive on all tested cell lines in 3D assay except SK-BR-3 cell line where moderate activity was still observed. The new insight into SAR guided by results from 3D screening showed us that translocation of 2-imidazolinyl group from benzimidazole ring to phenyl ring was detrimental for activity on three cell lines. Finally, importance of additional nitrogen found in structures 11a and 11b was proved in both assays. Although some slightly disagreements in sensitivity were observed on all tested cell lines, compounds 11a and 11b were active on both assays and this makes them a good candidate for further profiling.
Conclusions
To test the hypothesis of how differences in the activities of chemotherapeutic drugs in both 2D and 3D assays correlate with SAR data, we compared the drug sensitivities of the three human breast cancer cell lines (SK-BR-3, MDA-MB-231, T-47D) and two pancreatic cancer cell lines (MIA PaCa-2 and PANC-1) grown in 2D and 3D cell culture conditions. Doxorubicin, docetaxel, vinorelbine and gemcitabine were used as standard drugs and we compare these to our benzoimidazole-like compounds. Large variations in responses were observed between 2D and 3D models of standard drugs in comparison with variations in responses of tested compounds. This indicated that activity is dependent not only on structure similarity, but also on mechanism of action and cell culture susceptibility. SAR in 2D and 3D assays () reveals that activity of tested compounds is strongly depended on the rigidity of central core and nature of functional groups on benzimidazole or phenyl rings. Although the results for the tested compounds showed fewer variations between the 2D and 3D models, due to higher structural similarities and consequently more similar mode of action. However, Cyclic compounds (5a–5c, 7, 8, 10a–10c, 11a–11b) were generally more active comparing to acyclic (3a–3i, 4a–4d) systems. Further, the prerequisite of 2-imidazolinyl group was confirmed in several examples (3f–3g, 5a, 8, 11a–11b). Results obtained in 2D and 3D models are, in most cases, highly comparable but in some cases we have observed significant disagreement. This indicates that how some compounds can be discarded in an early phase of research, while chance was given to false positive compounds. To be sure which of cell culture systems is more accurate, further in vivo profiling is needed.
Declaration of interest
We greatly appreciate the financial support of the Croatian Ministry of Science Education and Sports under the project 125-0982464-1356 and Croatian Science Foundation under the project 5596 (Synthesis and cytostatic evaluations of novel nitrogen heterocycles library).
References
- Kim JB. Three-dimensional tissue culture models in cancer biology. Semin Cancer Biol 2005;15:365–77
- Siolas D, Hannon GJ. Patient-derived tumor xenografts: transforming clinical samples into mouse models. Cancer Res 2013;73:5315–19
- Abbott A. Cell culture: biology's new dimension. Nature 2003;424:870–2
- Schmeichel KL, Bissell MJ. Modeling tissue-specific signaling and organ function in three dimensions. J Cell Sci 2003;116:2377–88
- Breslin S, O’Driscoll L. Three-dimensional cell culture: the missing link in drug discovery. Drug Discov Today 2013;18:240–9
- Tung YC, Hsiao AY, Allen SG, et al. High-throughput 3D spheroid culture and drug testing using a 384 hanging drop array. Analyst 2011;136:473–8
- Hranjec M, Kralj M, Piantanida I, et al. Novel cyano- and amidino-substituted derivatives of styryl-2-benzimidazoles and benzimidazo[1,2-a]quinolines. Synthesis, photochemical synthesis, DNA binding and antitumor evaluation, Part 3. J Med Chem 2007;50:5696–711
- Hranjec M, Piantanida I, Kralj M, et al. Novel amidino-substituted thienyl- and furyl-vinyl-benzimidazole derivatives and their photochemical conversion into corresponding diaza-cyclopenta[c]fluorenes. Synthesis, interactions with DNA and RNA and antitumor evaluation, Part 4. J Med Chem 2008;51:4899–910
- Hranjec M, Lučić B, Ratkaj I, Karminski-Zamola G. Novel imidazo[4,5-b]pyridine and triaza-benzo[c]fluorene derivatives: synthesis, antiproliferative activity and DNA binding studies. Eur J Med Chem 2011;46:2748
- Sedić M, Poznić M, Gehrig P, et al. Differential anti-proliferative mechanisms of novel derivative of benzimidazo[1,2-a] quinoline in colon cancer cells depending on their p53 status. Mol Cancer Ther 2008;7:2121–32
- Weaver VM, Lelièvre S, Lakins JN, et al. β4 integrin-dependent formation of polarized three-dimensional architecture confers resistance to apoptosis in normal and malignant mammary epithelium. Cancer Cell 2002;2:205–16
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 1983;65:55–63
- Réjiba S, Bigand CH, Parmentier C, Hajri A. Gemcitabine-based chemogene therapy for pancreatic cancer using Ad-dCK::UMK GDEPT and TS/RR siRNA strategies. Neoplasia 2009;11:637–50
- Coyne CP, Jones T, Bear R. Anti-neoplastic cytotoxicity of gemcitabine-(C4-amide)-[anti-HER2/neu] in combination with Griseofulvin against chemotherapeutic-resistant mammary adenocarcinoma (SKBr-3). Med Chem 2013;3:210–23
- Awasthi N, Zhang C, Schwarz AM, et al. Comparative benefits of Nab-paclitaxel over gemcitabine or polysorbate-based docetaxel in experimental pancreatic cancer. Carciongenesis 2013;34:2361–9