Abstract
Human serum paraoxonase 1 (PON1, EC 3.1.1.2) is a high density lipoprotein (HDL)-associated antioxidant enzyme that not only decreases oxidative stress, but it is also implicated in development of many cancers. Genetic information provides a means of identifying people who have an increased risk of cancer, thus this knowledge of cancer genetics helps to identify the ability to characterize malignancies leading to the development of new therapeutic approaches. Because of this reason, in this preliminary study we aimed to investigate the role of human serum PON1 enzyme activity and phenotypic distribution in 32 breast cancer (BC) patients (age range 28–82) and 35 cancer free (CF) control group (age range 21–67). PON1 enzyme was prepared from the serum pool of BC patients using hydrophobic interaction chromatography on L-tyrosine-9-aminophenanthrene-coupled Sepharose 4Bgel. The PON1 enzyme activity towards paraoxon substrate was quantified spectrophotometrically. The basal activity of PON1 was statistically decreased in cancer cases compared to the control group. In addition, individuals were classified according to phenotyping of human PON1 Q and R types. In the cohort of BC patients, an increase in the frequency of the PON homozygote Q (AA) genotype was observed (31% in the BC group versus 14% in the CF controls). The frequency of the PON heterozygote QR (AB) genotype was 34.5% in the patients with BC and 37% in the CF group. The same trend was observed in PON homozygote R (BB) genotype frequency (BC cases 34.5% versus controls 49%). We determined that the kinetic parameters of the purified enzyme by Lineweaver–Burk method. We obtained Km and Vmax values of 0.227 mM and 62 U/mL min for the BC enzyme, compared with 0.775 mM and 206 U/mL min for the CF control enzyme. As a conclusion, it is clear from our results that while the PON1 AA allele frequency in BC cases is much higher, that of BB allele is much lower, in comparison with the control group. The most significant finding of this study is AA allele activity which is low in BC cases was found high. We concluded that decreased AA allele PON1 activity might have a relation with BC.
Introduction
The paraoxonase (PON) gene family, located on human chromosome 7q21.3–22.1, codes for a multifactorial antioxidant enzyme (EC 3.1.1.2)Citation1. The PON protein family is comprised of PON1, PON2 and PON3. PON1 is mostly synthesized in the liver and associates with high density lipoproteins (HDLs) in serumCitation2,Citation3. One of the best known HDL functions is the transport of plasma lipid hydroperoxide. HDLs are also carriers of the lipid hydroperoxide-destroying enzymes PON1, PON3 and glutathione phospholipid peroxidaseCitation4. These enzymes remove lipid peroxidation products. Lipid peroxidation causes oxidative stress in organisms. It is well known that oxidative stress is associated with cancer development and progressionCitation5.
The PON1 gene has promoter and coding region polymorphisms that affect gene expression levelsCitation6. Human serum PON1 exhibits two genetic polymorphisms. These result from exchange of the amino acids at the 55 or 192 positions. Either glutamine (Q allele) or arginine (L allele) can be at the 192 position and methionine (M allele) or leucine (L allele) at the 55 position. Both Q and R isoforms prevent oxidation of low density lipoproteins (LDLs). However, the R isoform possesses a higher affinity for paraoxon than the Q isoformCitation7. These polymorphisms are related with cancer development. Oxidative stress can damage biological molecules such as DNA and thus can lead to tumorogenesis. PON1 prevents oxidation of LDLs and detoxifies oxidative stress molecules that are carcinogenicCitation1,8–10.
Breast cancer (BC) is a type of cancer that predominantly influences the morbidity and mortality of females. Genetic factors and carcinogens are important contributors to the etiology of BC. Despite this, the molecular mechanisms involved in BC development are still unclearCitation10. Several studies have shown that mutations of oncogenes and tumor suppressor genes such as BRCA1 cause oxidative stress and cancer developmentCitation11. Some lipid peroxidation products also play a role in oncogenesisCitation12. Oxidized LDL is a casual factor for oxidative stress-related cancersCitation13. Conversely, HDL acts as an anticarcinogenic agent, as it provides enzymatic and non-enzymatic generation of reactive oxygen speciesCitation14.
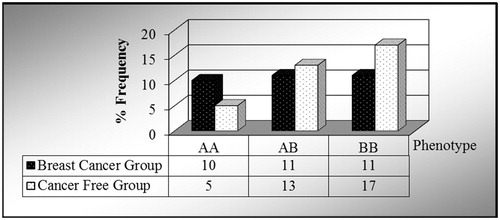
In this preliminary study, we aimed to investigate the PON1 enzyme in BC cases. It was found that the basal activity of PON1 was statistically decreased compared to the control group. Also, considering the important role of polymorphism in genetic susceptibility, we also determined that the phenotypic distribution in the cancer patients and control groups was AA 14%, AB 37% and BB 49%.
Materials and methods
Sepharose-4B, L-tyrosine, 9-aminophenanthrene, paraoxon, protein assay reagents and chemicals for electrophoresis were all obtained from Sigma Chem. Co. (Milan/Italy). All other chemicals used were of analytical grade and obtained from Sigma Chem. Co (Darmstadt/Germany). The study protocol was approved by the local human ethics body and informed consent was obtained from each patient. In total, 32 cases (age range 28–82) at different stages of BC from the Ali Osman Sönmez Oncology Hospital, Bursa/Turkey, were compared with 35 cancer free (CF), age-matched controls (age range 21–67). Statistical analyses were performed according to the HDL, LDL and cholesterol parameters between the BC and CF groups.
Phenotyping of human PON1 Q and R types
In order to classify individual phenotypes, two parameters were used. According to Eckerson et al.Citation15, the phenotypic distribution of PON1 activity is determined by the ratio of basal PON activity and PON activity in the presence of 1 M NaCl. After the basal and salt activity measurements of paraoxonase enzyme, phenotype determination is made by the formula given below:
Basal paraoxonase activity
For the activity measurement, 0.05 mL serum sample is immediately added in 1 mL buffer (100 mM Tris–base pH: 10.5) and substrate solution (1 mM paraoxon). Following this addition, basal activity occurred in absorption is measured for 1 min at 412 nm 37 °C. Later, coenzyme (1 M NaCl) is added to the same solutions and salt stimulate activity is measured. By this way, the enzymatic conversion rate of paraoxonto p-nitrophenol is detected. The same procedure is repeated without enzyme and the difference between these two is determined as the activity of enzyme. One unit paraoxonase is accepted as p-nitrophenol which occurred in one minute, μmol.
Individuals were classified for paraoxonase phenotype using the antimode at 60% stimulation as the dividing point between the nonsalt-stimulated, Q type, and the salt-stimulated, QR (60–200%) and R (200%-up) types.
Purification of human PON1
Human serum was isolated from fresh human blood collected in a dry tube. The blood samples were centrifuged at 26 916 g for 15 min and the serum was recovered. Firstly, crude serum PON1 was isolated by ammoniumsulfate precipitation (60–80% fraction)Citation16. The precipitate was collected by centrifugation at 26 916 g for 20 min and then redissolved in 100 mM Tris–HCl buffer (pH 8.0)Citation17.
The crude PON1 solution was then subjected to hydrophobic interaction chromatography on a hydrophobic column of Sepharose-4B-coupled L-tyrosine-9-aminophenanthrene, synthesized according to Gençer et al.Citation17. Briefly, a 1:1 suspension of Sepharose-4B in water was activated by treatment with 10% CNBr. The mixture was titrated to pH 11 in an ice bath and maintained at that pH for 8–10 min. The reaction was stopped by filtering the gel on a Buchner funnel and washing with cold 0.1 M NaHCO3 buffer, pH 10.0. L-tyrosine was then coupled to the activated Sepharose-4B by addition of a saturated L-tyrosine solution in 0.1 M NaHCO3 buffer of pH 10.0. The reaction was completed by stirring with a magnet for 90 min. The L-tyrosine–Sepharose-4B gel was then extensively washed with distilled water to remove the excess of L-tyrosine. The final hydrophobic gel was obtained by diazotization of 9-aminophenanthrene and coupling of this compound to the L-tyrosine–Sepharose-4B gel. The pH was adjusted to 9.5 with 1 M NaOH and, after gentle stirring for 3 h at room temperature, the coupled Red Sepharose derivative was washed with water and then an excess of 0.05 M Tris–sulfate, pH 7.5. The column was finally equilibrated with 0.1 M Na2HPO4 buffer, pH 8.0 containing 1 M ammonium sulfate.
The paraoxonase was eluted with ammonium sulfate gradient using 0.1 M Na2HPO4 buffer with and without ammonium sulfate pH 8.00. The purified PON enzyme was stored in the presence of 2 mM CaCl2 at +4 °C, in order to maintain activityCitation17.
Paraoxonase enzyme assay
PON1 enzyme activity towards paraoxon substrate was quantified spectrophotometrically by the method described by Gan et al.Citation18. The reaction was followed for 2 min at 37 °C by monitoring the appearance of p-nitrophenol at 412 nm in a Biotek automated recording spectrophotometer. A molar extinction coefficient (ɛ) of 17 100 M−1 cm−1 for p-nitrophenol in 100 mM Tris–base buffer, pH 8.0 was used for the calculation. PON1 activity (1 Ul−1) was defined as 1 μmol of p-nitrophenol formed per minute.
Statistical analysis
Statistical analyses were performed using the SPSS software package, version 10.0 for Windows. Clinical laboratory data were expressed as mean ± standard deviation. Mean values were compared between patients with BC and healthy individuals by Student's t test.
Results and discussion
PON1 was purified from two separate pooled serum samples, obtained from BC patients and control individuals, respectively, by a two-stage process involving ammonium sulfate fractionation followed by hydrophobic interaction chromatography on a prepared 9-aminophenanthrene-L-tyrosine–Sepharose4BCitation17. The purified PON1 gave a single band of 43 kDa molecular weight on an SDS-PAGE gel.
Our subsequent study has two main parts: firstly a comparative analysis of the polymorphism of PON1 in BC cases; and secondly kinetic studies of PON1 from both BC and (CF) groups.
In the first part, we investigated PON1 enzyme polymorphism in 32 BC cases compared to 35 CF control serum samples. In our BC test group, we found that the frequency of the AA phenotype was 31%, AB was 34.5% and BB was 34.5%. In contrast, the CF control group gave frequencies of 14%, 37% and 49%, respectively. It is thus clear from our results, that while the PON1 AA allele frequency is much higher, that of the BB allele is much lower, in comparison with the control group (). So there is no significant relation between BC and control group.
In the second part of the study, we combined serum samples from the 32 BC patients (aged between 28 and 82, and in different stages of BC) to give a single serum pool. Likewise, we combined serum samples from the CF control group of 35 women (aged 21–67 and who have not been diagnosed with any serious illnesses). PON1 was then purified by hydrophobic interaction chromatography from each of the two pools, and the kinetic constants were measured and compared between pools. We obtained Km and Vmax values of 0.227 mM and 62 U/mL min for the BC enzyme, compared with 0.775 mM and 206 U/mL min for the CF control enzyme.
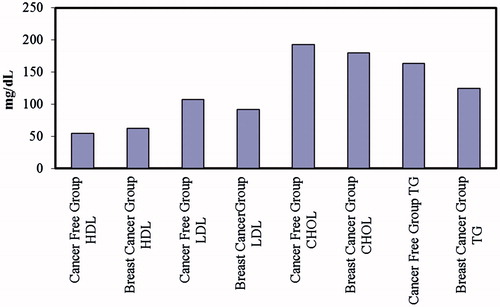
In addition, we also compared measurements of various lipid parameters, i.e. cholesterol, triglyceride, HDL and LDL (mg/dL) values, with the basal PON1 activity, and also PON1 activity induced by high salt, for both CF and BC groups ( and ). Phenotype determination was also performed, using the ratios of PON1 basal activities and PON1 activities induced with salt. In the control group, the basal activity was generally between 0 and 50 U, though some values >50 U values were also observed. After salt induction, values of 40–200 U were generally obtained, with a few cases being above or below this range. Cholesterol values were 150–250 mg/dL, triglyceride 50–150 mg/dL, HDL 30–70 mg/dL, and LDL between 50 and 200 mg/dL. In the BC group, the basal PON1 activity was between 15 and 70 U, which after salt induction was raised to 25–190 U. Lipid measurements gave values of cholesterol as 115–240 mg/dL, triglyceride 50–235 mg/dL, HDL 30–90 mg/dL and LDL 30–160 mg/dL ( and ).
Figure 2. The graph of basal activities of PON 1 and salt stimulated PON1 activities of BC group and CF group.
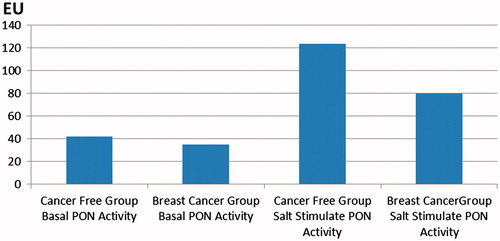
Table 1. Serum paraoxonase enzyme activities, lipid and other parameters of BC cases.
Table 2. Serum paraoxonase enzyme activities, lipid and other parameters of CF group.
Many studies have investigated the links between common genetic variants and cancer risk, as most common cancer types are influenced by population and family genetics in which shared genetic variations contribute a proportion of cancer riskCitation19. There are several studies about the putative relationship between PON polymorphism and cancerCitation1,Citation2,Citation8,Citation19. However, our study is the first attempt to investigate PON1 polymorphism in Turkish female BC patients. It is clear from the literature that PON1 enzyme activity varies according to polymorphism. One of the most important results of this study relates to the AA allele of PON1, which has a lower enzymatic activity and occurs with a much higher frequency in BC patients. Indeed, many researchers have suggested that low PON1 activity may increase cancer riskCitation20,Citation21. Despite its physiological and therapeutic importance, the structure and mechanism of action of PON1 have yet to be elucidated.
Controversy results have been obtained in the majority of studies about PON gene polymorphisms and risk of BC. Saadat had identified six eligible studies, including 3943 subjects (1608 patients and 2335 healthy controls) in relation to the polymorphisms of PON1 and risk of BCCitation1 and he had found from the meta-analysis of published data, M and Q alleles of the PON1 gene were associated with an increased risk of BC according to the comparative studies from Italy, Egypt, USA, Malaysia and Turkey. However, another study, investigating BRCA1 mutations in BC, demonstrated an association of the Q192R and L55M PON1 SNPs with BC. According to Steven’s group, both of these polymorphisms are common and influence PON1 activityCitation20, and these results are the most positive in suggesting that PON1 may influence BC development.
In another study done in 100 BC, Egyptian females and this study has confirmed the M allele of PON1 L55M polymorphism could be a suitable marker for BC susceptibility and tumor prognosis in Egyptian womenCitation21. Also Ağaçhan et al. had investigated the risk factor of BC relation of PON 1 and they had found PON1 192 genotype distribution have different risk ratios between BC and control groupCitation10. According to their results they had confirmed the relation between oxidative stress and BC risk in Turkish females.
In our preliminary study, the sample of this research consists of thirty three 28–82-year-old women diagnosed with BC and 21–67-year-old 30 healthy women. The enzyme polymorphism of the PON1 serum found in the blood taken from the sample was studied. While the AA phenotype was 31%, ABB phenotype was 34.5% and BB phenotype was 34.5% in the experimental group; the values were respectively found as 14%, 37%, 49% in the control group. The results indicate that PON1 AA allele is higher in the experimental group whereas the BB allele is lower than the one in the control group (). The most significant finding of this study is the fact that AA allele activity which is low in BC cases is found high.
Previous studies (carried out so far) have shown that more than 160 polymorphisms are detected in PON1 gene’s coding sequences, introns and genes regulatory part which is a promoterCitation22. Polymorphism in PON1 enzyme’s activity is different and varies from others in terms of ethnic originCitation23. At first, in 1973, Von Mallinckrodt and his coworkers demonstrated that PON 1 enzyme shows genetic polymorphisms and activities of enzyme have trimodal distributionCitation24. Molecular basis of paraoxonase activity polymorphism is related to the displacement of two amino acids by spontaneous mutations in coding sequence. One of these mutations in coding sequence is occurred due to the change from glutamine (Q) to arginine (R) in codon 192.
In addition to PON1, M/L55 and Q/R192 polymorphisms in the coding sequence of PON1, at least five more polymorphisms are detected on promoter sequence. These polymorphisms are found in −107/−108 (C/T), −126 (C/G), −160/−162 (A/G), −824/−832 (A/G) and −907/−909 (C/G). Polymorphisms in the promoter of PON1 gene have great effects on gene expression and serum levels of enzymes. Polymorphisms in promoter are also found in PON1 gene 3′ non-coding sequence but the importance of them is not known yet.
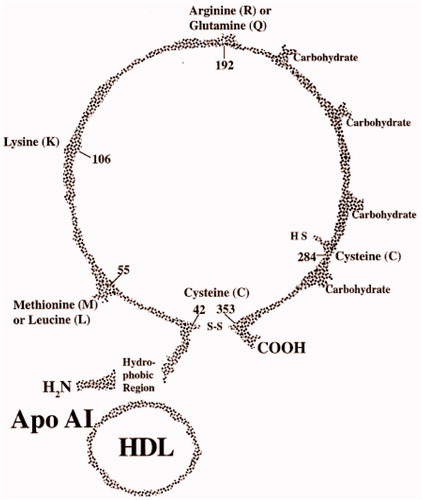
Paraoxonase polymorphic distribution shows considerable alterations between the races. In Turkish population, RR allele was found at quite low levels. For tri modal distribution, the frequency of QQ, QR and RR alleles were detected as 48.6%, 41.0% and 10.4%Citation25. Q allele which shows low activity is high in Europe, Canada, America and low in Australia, Aborigine and Zambia. PON1 gene shows two important polymorphisms which are Q/R 192 and M/L 55. The most common one is Q/R 192 polymorphism. Because the polymorphism in PON1 enzyme activity is dependent on substrate and it varies from population to population which has different ethnic origins, this was shown by the studiesCitation26. The molecular basis of paraoxonase activity polymorphism is related to the displacement of two amino acids by spontaneous mutations in coding sequence. One of these mutations in coding sequence is occurred by the change from glutamine (Q) to arginine (R) in codon 192Citation27. This polymorphism in PON1 activity is dependent on substrate. Second change in coding sequence is the conversion of leucine (L) to methionine (M) located on 55th position ()Citation28. It is found that this mutation has low effect on PON1 activity whereas it affects the level of protein in serumCitation29,Citation30. Paraoxon hydrolysis activity of protein that is coded by R allele of PON1 is eight times higher when it is compared to Q alleleCitation31. Also, Q form of PON1 is more effective in oxidized HDL and LDL metabolism compared to R formCitation32. This genetic polymorphism also affects the concentration of serum protein. Homozygote R individuals have high enzyme concentration when it is compared to homozygote Q individualsCitation33. Polymorphic distribution in different populations causes variation between individualsCitation34. Phenotyping methods are developed by the discovery of qualitative and quantitative difference in allozymes of PON1. At first, in 1983, two phenotypes were defined by Eckerson which is based upon different responses of two isoenzymes to salt and pHCitation15. In the presence of salt (NaCl), homozygous R allozyme (RR) has high enzyme activity to paraoxon.
Figure 4. The gene polymorphisms of PON1Citation28.
In our second part of the study, we used hydrophobic interaction chromatography to purify PON1. One of the most important reasons for the selection of this technique is the known hydrophobic character of PON1Citation35. The H1 hydrophobic sequence between residues 7 and 18 includes leucine, phenylalanine, proline, isoleucine, tyrosine, tryptophane and valine residues. The H2 hydrophobic sequence occurs between residues 185 and 202. In addition, there is a partially hydrophilic region between PON1’s hydrophobic surface and HDL. This region is rich with tryptophane, tyrosine and lysine amino acids. Sinan and colleagues purified human serum PON1 and determined the Km and Vmax values of the enzyme to be 4.16 mM and 227.27 EU, respectivelyCitation16. Kılınç and his group purified the human 192Q and 192R PON1 isoenzymes using a multi-step procedure. They measured the kinetic values of 192Q and 192R PON1 for homocysteine thiolactone, which were 23.5 mM and 22.6 mM, respectively. For 192R PON1, the Vmax was 2.5-fold higher and the kcat/Km was 2.6-fold higher than the respective values for the 192Q PON1, using homocysteine thiolactone as the substrateCitation36.
Our study is the first report for the purification of PON1, by a rapid one step procedure with high yield, from serum pools derived from both BC and control groups. We have then compared data on both enzyme kinetics and PON1 polymorphisms, which have revealed differences in both parameters and we obtained Km and Vmax values of 0.227 mM and 62 U/mL min for the BC enzyme, compared with 0.775 mM and 206 U/mL min for the CF control enzyme. Therefore, according to our results, the most significant finding of this study is the fact that AA allele activity which is low in BC cases is found high, in fact literature review also shows that the low PON1 activity increases the cancer risk. The future challenges may identify novel pathways according to the enzyme kinetics which provide new targets for therapeutic intervention in the practice of clinical medicine.
Acknowledgements
The authors thank Dr Erol Aksaz (Ali Osman Sonmez Oncology Hospital Bursa/Turkey) for advice and for providing the blood samples for this study. The authors are also thankful to Dr Malcolm Lyon for his invaluable contribution on this paper.
Declaration of interest
This study was supported by the Balıkesir University Scientific Research Foundation (BAP 2008/31).
The authors do not declare any conflict of interest relating to this study.
References
- Saadat M. Paraoxonase 1 genetic polymorphisms and susceptibility to breast cancer: a meta-analysis. Cancer Epidemiol 2012;36:101–3
- Naidu R, Har YC, Taib NA. Genetic polymorphisms of paraoxonase 1 (PON1) gene: association between L55M or Q192R with breast cancer risk and clinico-pathological parameters. Pathol Oncol Res 2010;16:533–40
- Mackness B, Durrington PN, Mackness MI. Human serum paraoxonase. Gen Pharmacol 1998;31:329–36
- Barter PJ, Nicholls S, Rye KA, et al. Antiinflammatory properties of HDL. Circ Res 2004;95:764–72
- Balci H, Genc H, Papila C, et al. Serum lipid hydroperoxide levels and paraoxonase activity in patients with lung, breast, and colorectal cancer. J Clin Lab Anal 2012;26:155–60
- Hofer SE, Bennetts B, Chan AK, et al. Association between PON1 polymorphisms, PON activity and diabetes complications. J Diabetes Complications 2006;20:322–8
- Aldırmaz M, Altıntaş N, Var A, Ellidokuz E. Role of the PON polymorphisms on progression of chronic hepatitis and cirrhosis. Turk J Biochem 2011;36:255–60
- Fang DH, Fan CH, Ji Q, et al. Differential effects of paraoxonase 1 (PON1) polymorphisms on cancer risk: evidence from 25 published studies. Mol Biol Rep 2012;39:6801–9
- Liu C, Liu L. Polymorphisms in three obesity-related genes (LEP, LEPR, and PON1) and breast cancer risk: a meta-analysis. Tumour Biol 2011;32:1233–40
- Ağaçhan B, Yaylım I, Ergen HA, et al. Is paraoxonase1 192 BB genotype risk factor for breast cancer? Adv Mol Med 2006;2:37–40
- Vurusaner B, Poli G, Basaga H. Tumor suppressor genes and ROS: complex networks of interactions. Free Radic Biol Med 2012;52:7–18
- Elkiran ET, Mar N, Aygen B, et al. Serum paraoxonase and arylesterase activities in patients with lung cancer in a Turkish population. BMC Cancer 2007;7:48–15
- Barrera G. Oxidative stress and lipid peroxidation products in cancer progression and therapy. ISRN Oncol 2012;2012:137289
- Chander R, Kapoor NK. High-density lipoprotein is a scavenger of superoxide anions. Biochem Pharmacol 1990;40:1663–5
- Eckerson HW, Wyte CM, La Du BN. The human serum paraoxonase/arylesterase polymorphism. Am J Hum Genet 1983;35:1126–38
- Sinan S, Kockar F, Arslan O. Novel purification strategy for human PON1 and inhibition of the activity by cephalosporin and aminoglikozide derived antibiotics. Biochimie 2006;88:565–74
- Gencer N, Arslan O. Purification human PON1Q192 and PON1R192 isoenzymes by hydrophobic interaction chromatography and investigation of the inhibition by metals. J Chromatogr B Analyt Technol Biomed Life Sci 2009;877:134–40
- Gan KN, Smolen A, Eskerson HW, La Du BN. Purification of human serum paraoxonase/arylesterase. Evidence for one esterase catalyzing both activities. Drug Metab Dispos 1991;19:100–6
- Dunning AM, Healey CS, Pharoah PD, et al. A systematic review of genetic polymorphisms and breast cancer risk. Cancer Epidemiol Biomarkers Prev 1999;8:843–54
- Stevens VL, Rodriguez C, Pavluck AL, et al. Association of polymorphisms in the paraoxonase 1 gene with breast cancer incidence in the CPS-II Nutrition Cohort Cancer. Epidemiol Biomarkers Prev 2006;15:1226–8
- Hussein YM, Gharib AF, Etewa RL, ElSawy WH. Association of L55M and Q192R polymorphisms in paraoxonase 1 (PON1) gene with breast cancer risk and their clinical significance. Mol Cell Biochem 2011;351:117–23
- Costa LG, Cole TB, Jarvik GP, Furlong EF. Functional genomics of the paraoxonase (PON1) polymorphisms: effects on pesticide sensitivty, cardiovascular disease, and drug metabolism. Annu Rev Med 2003;54:371–92
- Costa LG, Furlong CE. Paraoxonase (PON1) in health and disease: basic and clinical aspects. Norwell (MA): Kluwer Academic Publishers; 2002
- Geldmacher-von Mallinckrodt M, Lindorf HH, Petenyi M, et al. Genetically determined polymorphism of human serum paraoxonase (E.C.3.1.1.2). Humangenetic 1973;17:331–5
- Aynacıoğlu AS, Kepekçi Y. The human paraoxonase Gln-Arg 192 (Q/R) polimorphism in Turkish patients with coronary artery disease. Int J Cardiol 2000;74:33–7
- Marchesani M, Hakkarainen A, Tuomainen TP, et al. New paraoxonase 1 polymorphism I102V and the risk of prostate cancer in Finnish men. J Natl Cancer Inst 2003;95:812–18
- Humbert R, Adler DA, Disteche CM, et al. The molecular basis of the human serum paraoxonase activity polymorphism. Nat Genet 1993;3:73–6
- Juretic D, Tadijanovic M, Rekic B, et al. Serum paraoxonase activities in hemodialyzed uremic patients: cohort study. Clin Sci 2001;42:146–50
- Brophy VH, Hastings MD, Clendenning JB, et al. Polymorphisms in the human paraoxonase (PON1) promoter. Pharmacogenetics 2001;11:77–84
- Brophy VH, Jampsa RL, Clendenning JB, et al. Effects of 5′ regulatory-region polymorphisms on paraoxonase-gene (PON1) expression. Am J Hum Genet 2001;68:1428–36
- Davies HG, Richter RJ, Keifer M, et al. The effect of the human serum paraoxonase polymorphism is reversed with diazoxon, soman and sarin. Nat Genet 1996;4:334–6
- Aviram M, Hardak E, Vaya J, et al. Human serum paraoxonases (PON1) Q and R selectively decrease lipid peroxides in human coronary and carotid atherosclerotic lesions: PON1 esterase and peroxidase-like activities. Circulation 2000;101:2510–17
- Mackness MI, Mackness B, Durringhton PN, et al. Paraoxonase: biochemistry, genetics and relationship to plasma lipoproteins. Curr Opin Lipid 1996;7:69–76
- Ginsberg G, Neafsey P, Hattis D, et al. Genetic polymorphism in paraoxonase 1 (PON1): Population distribution of PON1 activity. Toxicol Environ Health B Crit Rev 2009;12:473–507
- Salman M, Malleda C, Suneel NA, et al. Homology modeling of human serum paraoxonase1 and its molecular interaction studies with aspirin and cefazolin. Bioinformation 2011;7:59–63
- Bayrak A, Bayrak T, Demirpençe E, Kılınç K. Differential hydrolysis of homocysteine thiolactone by purified human serum (192)Q and (192)R PON1 isoenzymes. J Chromatogr B Analyt Technol Biomed Life Sci 2011;879:49–55