Abstract
The finding of the most appropriate way to assess precisely the antivenom efficacy represents one of the major issues for antivenom standardization and success increasing of antivenom therapy. The efficacy of experimental Vipera ammodytes antivenom raised in sheep was determined using in vivo mouse lethality test, respectively, L-aminoacid oxidase, total proteinase and phospholipase A2 antienzymatic effectiveness. The values gained for the antivenom potency depend on the method of measure. So, some of the most toxic venom proteins own phospholipase A2 activity and provide the highest antivenom potency (lowest effective dose) values by antienzymatic assay method. This value is similar with total antiproteolytic antivenom potency value, but almost three times higher than value obtained by L-aminoacid oxidase (low toxic viper venom protein) antienzymatic assay method.
Introduction
The antivenoms are the most efficient pharmaceutical tools against the snakes bite accidentsCitation1–3. This kind of medicines is obtained from the hyperimmunized animals with one or more snake species venomsCitation4–6. If the old antivenoms were separated and used as crude serum, the modern antivenoms embody mainly purified immunoglobulins concentratesCitation7,Citation8.
Many factors affect the antivenom therapy, but the standardization and subsequently the methods to assess the product efficacy are very important. The antivenom capability to annihilate certain toxic or biochemical venom properties grants the antivenom potency and can be expressed as median effective dose, ED50Citation1,Citation9.
One of the most used and aged antivenom potency assessing method is based on the injection in the animal experiment bodies of a significant venom–antivenom mixture and interpolation of the 50% survival animalCitation1,3,7,8,10,11. Another way is represented by immunoenzymatic method for the assay of antivenom potencyCitation11–14. In these cases, antivenom potency is directly articulated on venom unit quantity but, as it is already well known, even the venom of the same snake species differs (sometime substantially) in theirs biological and toxicological properties. Consequently, the antivenom efficiency based on the assay of the specific venom biological properties will complete very well the whole story of antivenom potency.
Because all venoms (particularly harvested from snake) possess a very significant enzymatic content, the antivenom potency can be also determined by antienzymatic assayCitation1,Citation13,Citation15,Citation16. The antivenom efficacy values obtained by all previously mentioned methods can be indirectly compared using correlation coefficientsCitation15,Citation16. The results of four potency assay methods for an experimental ovine immunoglobulinic concentrate are presented below.
Methods
All reagents used in this study have analytical grade purity. Crude dried Vipera ammodytes venom was purchased from the local herpetologist and extensively biochemically analysed.
The antivenom was obtained as immunoglobulins concentrate by caprylic acid precipitation and dialisys methodCitation8, from sheep serum (Ovis aries, 2 years old males), previously hyperimmunized with Vipera ammodytes venom.
In order to assay the antivenom potency, two stages were performed: first, certain venom–antivenom incubation mixtures were tempered at 37 °C for 30 min, and finally, the enzyme activities or survivor mice were evaluated. The nonlinear regression algorithms were used for experimental data management. Based on the best fitting curve equations, the ED50 s values were calculated.
In case of in vivo lethality tests, the ED50 correspond to the 50% surviving mice incubation ratio, but for the antienzymatic assay tests, we assume that ED50 match to the 50% enzyme activity decreasing. This approach let us to compare directly the antivenom potency value obtained by “in vivo” and “in vitro” methods. The antivenom effectiveness is expressed as milligrams of crude-dried venom with 50% of mouse lethality or enzymes activities neutralized by 1 mL of antivenom.
L-Amino-acid-oxidase antienzymatic potency
We determined the L-amino-acid-oxidase activity by peroxidase–hydrogen peroxide assay methodCitation17,Citation18. The reagents were tempered at 25 °C for 45 min.
About 0.5 mL solution of 1 mg/mL crude-dried venom in 0.9% NaCl was mixed with a volume of antivenom with a value of the following: 0.0, 0.1, 0.2, 0.3, 0.4 mL. The mixture was brought to the volume of 1 mL using Tris buffer solution, 0.2 M, pH 7.6 and incubated at 37 °C for 30 min.
About 0.1 mL of the previous incubation mixture was mixed with 0.1 mL peroxidase solution (approximately. 100 I.U./mL) and 1.8 mL Tris buffer solution, 0.2 M, pH 7.6, containing 10 mM 4-aminoantipyrine, 30 mM phenol and 3 mM L-phenylalanine. The absorbance of sample versus blank at λ = 507 nm was measured after 20 min in 1 cm width cell and the result was interpolated from the calibration curve previously obtained, using standard solutions of L-phenylalanine and long reaction time (approximately 3–4 h) or standard solutions of H2O2.
The (apparent) enzymatic activity of each incubation mixture sample was calculated as 0.4 absorbance (units) and expressed in micromoles aminoacid/min/mg venom. All reagent solutions used for this determination were prepared with Tris buffer solution, 0.2 M, pH 7.6.
Phospholipase A2 antienzymatic potency
We determined the phospholipase activity by a fatty acids pH-stat titration methodCitation19. The reagents were tempered at 25 °C for 45 min. About 0.5 mL solution of 2 mg/mL crude-dried venom in 0.9% NaCl was mixed with a volume of antivenom with a value of 0.0, 0.125, 0.25, 0.5 mL. The mixture was brought to the volume of 1 mL using Tris buffer solution, 0.02 M, pH 8.6 and incubated at 37 °C for 30 min.
Twenty-five per cent fresh egg yolk in distilled water was prepared with 0.15% CaCl2 and 0.2% sodium deoxicholate, the pH was brought to 8.6 with 0.2N NaOH solution (cca. 5 mL for 70 mL egg yolk solution) and tempered at 25 °C for 15 min. The pH was monitored and adjusted until remained stable (not decreasing with more than 0.02 units in 5 min).
About 0.05 mL incubation mixture (containing 0.050 mg solid venom) was added to the egg yolk solution and the time for pH-static titration (using ultratitrator or laboratory pH-meter) with 0.2N NaOH solution was measured.
The (apparent) enzymatic activity of each incubation mixture sample was calculated as 0.4 Volume (mL)/times (minutes) and expressed in micromoles fatty acids/min/mg venom. According to our data, the reaction is substrate zero-kinetic with 70 mL egg yolk 25% and 0.05 mg solid pure venom for maximum 7–9 min. We recommend reverse titration after adding 0.05–0.3 mL 0.2N NaOH solution, by time (minutes) measuring until pH reaches initial value.
Total proteolytic antienzymatic potency
We determined the proteolytic activity as caseinolytic activity by Folin–Ciocalteu tyrosine residues assay methodCitation20. The reagents were tempered at 25 °C for 45 min.
About 0.5 mL solution of 2 mg/mL crude dried venom in 0.9% NaCl was mixed with a volume of antivenom with a value of 0.0, 0.1, 0.2, 0.4 mL. The mixture was brought to the volume of 1 mL using Tris buffer solution, 0.4 M, pH 8.5 and incubated at 37 °C for 30 min.
About 0.5 mL 2% casein Tris buffer solution, 0.4 M, pH 8.5 and 0.1 mL venom–antivenom incubation mixture were tempered for 15 min at 37 °C. The reaction was stopped with 0.1 mL 0.4 M trichloroacetic solution. After 10 min, the sample was diluted with 0.4 mL distilled water and centrifugated for 5 min at 3000 rot/min.
About 0.5 mL supernatant and 2.5 mL 0.4 M sodium carbonate solution were treated with 0.5 mL Folin–Ciocilteu reagent and the absorbance of sample versus blank at λ = 660 nm was measured after 20 min in 1 cm width. The result was interpolated from the calibration curve, previously obtained using standard solutions of L-tyrosine. The (apparent) enzymatic activity of each incubation mixture sample was calculated as 0.39Absorbance (units) and expressed in micromoles Tyr/min/mg venom.
Mouse lethality assay potency
We determined the potency as effective median dose by intravenous mouse lethalityCitation1,Citation3,Citation9. The reagents were tempered at 25 °C for 45 min.
About 0.6 mL solution of 1 mg/mL crude-dried venom in 0.9% NaCl was mixed with a volume of antivenom with a value of: 0.0; 0.05; 0.1; 0.15; 0.2; 0.25 and 0.3 mL. The mixture was brought to the volume of 1 mL using 0.9% NaCl solution and incubated at 37 °C for 30 min. Seven groups of four 30 g NMRI mice male each were intravenously challenged with 0.2 mL incubation mixture containing 0.12 mg solid venom (corresponding to 5 LD50s) into tail vein. The survivor animals from each group were recorded after 48 h.
Results
The experimental data were represented in the corresponding graph and curve equation. The enzymatic activity values were plotted against the antivenom–venom incubation ratio and the best fitted curve were found by nonlinear regression algorithms. The ED50 values for each method are illustrated in .
Table 1. The experimental vipera ammodytes antivenom potency.
The antienzymatic potency assay curve equation for all studied venom enzymes has the same profile: y = (a + bx)/(1 + cx + dx2)where y = enzyme activity, x = antivenom/venom incubation ratio.
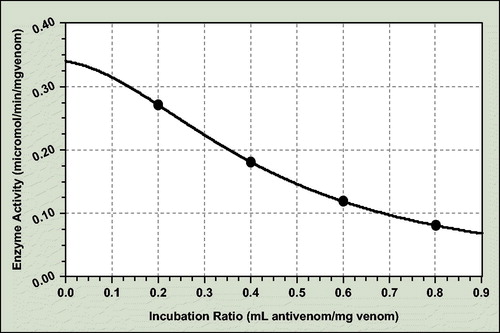
For the L-amino-acid-oxidase antienzymatic potency assay, the coefficient data are as follows ():
a = 0.340; b = 0.013; c = 0.378; d = 4.725
The incubation ratio for 50% neutralization of crude dried venom L-amino-acid-oxidase activity is 0.429 mL antivenom/mg venom and consequently ED50 = 2.33 mg venom/mL antivenom.
The enzyme activity of precisely ratio antivenom–venom incubation mixtures was determined spectrophotometricaly at 507 nm with peroxidase and Tris buffer solution, 0.2 M, pH 7.6, containing 10 mM 4-aminoantipyrine, 30 mM phenol and 3 mM L-phenylalanine.
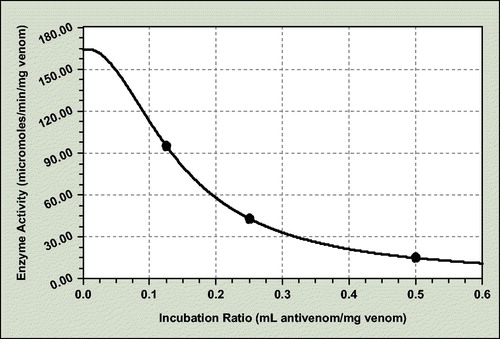
For the Phospholipase A2 antienzymatic potency assay, the coefficient data are as follows ():
a = 164; b = 126.991; c = 0.085; d = 56.495
The incubation ratio for 50% neutralization of crude-dried venom phospholipase A2 activity is 0.147 mL antivenom/mg venom and consequently ED50 = 6.80 mg venom/mL antivenom.
The enzyme activity of exactly ratio antivenom–venom incubation mixtures was determined pH-stat-titrimetricaly in 25% egg yolk solution with NaOH solution 0.2N.
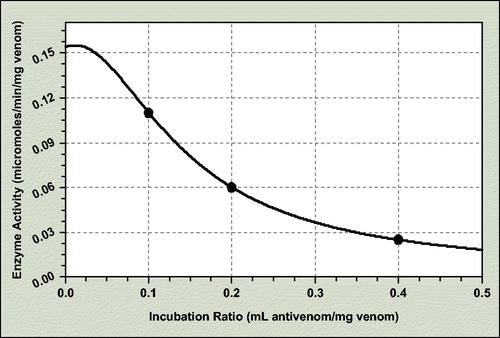
For the total protease antienzymatic potency assay, the coefficient data are as follows ():
a = 0.154; b = 0.317; c = 0.648; d = 62.392
The incubation ratio for 50% neutralization of crude-dried venom total proteinolytic activity is 0.157 mL antivenom/mg venom and consequently ED50 = 6.37 mg venom/mL antivenom.
The enzyme activity of precisely ratio antivenom–venom incubation mixtures was determined spectrophotometricaly at 660 nm with Folin–Ciocilteu reagent for L-tyrosine resulted after casein proteolysis.
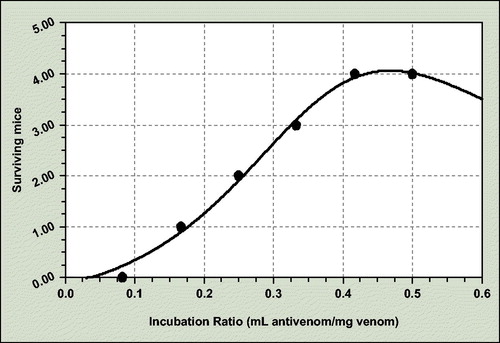
For the mouse lethality assay potency, the coefficient data are as follows () :
a = −0.135; b = 3.788; c = −3.493; d = 4.737
The incubation ratio for 50% neutralization of crude-dried venom mouse lethality is 0.256 mL antivenom/mg venom and consequently ED50 = 3.90 mg venom/mL antivenom.
The survivor mice number after challenged intravenously with 0.2 mL of various ratio antivenom/venom incubation mixtures was recorded and plotted against the antivenom/venom incubation ratio.
Discussions
It is generally agreed that certain type of venom enzymatic activity is due to more than one protein macromolecule category. As a result, when a venom enzymatic activity is measured in fact, the sum of the enzyme activity of all protein macromolecules with the same enzyme activity is measured. On the other hand, each venom enzyme macromolecule possesses several epitopes that generate, each, his own antibody population, each of them with his specific bound locus and inhibitory action mechanism.
If concept of inhibition is suitable to discuss interaction between one categories of enzyme with one inhibitor type, the simultaneous interaction between one enzyme and several types of inhibitors is much complicated, but not so complicate as simultaneous interaction between several enzyme (with the same type of activity) and several type of inhibitors for each of these enzymes. Consequently, when we study the interaction between venoms and antivenom using enzyme analysis instrument, it is very appropriate to substitute the basic concept of inhibition with antienzymatic perception. Resuming, due to the venom and antivenom constituents complexity, the antienzymatic effects integrate the various antibodies population inhibitory action upon different molecular enzyme surface epitopes. Moreover, in case of phospholipase, but especially in case of proteases, different venom proteins are involved in the same enzymatic activity typeCitation21. All this aspects are mathematically articulated by the equations revealed with nonlinear regression algorithm. Accordingly to the Michaelis–Menten theory, in the presence of the inhibitor enzyme velocity have form V = c1/(c2 + c3I), with V – enzyme velocity, I – inhibitor concentration, and c1, c2, c3 – constants, but for the two reaction pathway, the combined velocity will have the form like this: V = (k1+ k2I)/(1+ k3I + k4I2), with k1, k2, k3, k4 – constants. Because the venom concentrations and volumes for enzymatic reaction during of experimental condition are constant, it is easily to observe that enzymatic activity have the form identical with that previously revealed by nonlinear regression algorithm, y = (a + bx)/(1 + cx + dx2), where y = enzyme activity, x = incubation ratio. Based on this enzyme activity curve equations outline, some interesting broad-spectrum information can be extract. It is suggested that, mainly two epitope-populations antibody alter L-aminoacid-oxidase-active catalytic centre functionality. Much plausible, total measured proteinase activity owing mainly to the two enzymes, each one inhibited by one epitope-population antibody from the immunoglobulins concentrate. The situation is similar in case of phospholipase. We emphasize these deductions do not exclude the contribution of many other pathway for each enzyme activity but these are not significant.
Conclusions
For this experimental antivenom, the previous results allow conclusion that various ED50 values of antivenom potency obtained by antienzymatic assay seem to depend on the biological activities of the proteins involved in the enzymatic assay.
So, L-aminoacid-oxidase antienzymatic assay method provides the lowest ED50 value of antivenom potency, but phospholipase A2 antienzymatic assay method supplies the highest ED50 value of antivenom potency, similar with ED50 value obtained with total protease antienzymatic assay method. It is generally recognized that viperid venom phospholipases A2 and proteases produce the most damaging effects in the animal body and is unsurprisingly the strong immune response with elevated antibody concentration for these enzymes in the sheep blood serum. Compared with antienzymatic assay methods, “in vivo” mouse lethality test provides intermediate ED50 value of antivenom potency because the test animal body mediates the whole venom toxicity.
Due to precision and resources reasonability, the antienzymatic antivenom potency assay seems to become very suitable strategy for better pharmaceutical and biochemical characterization of antivenoms. This approach is very useful for antivenom standardization, including during immunoglobulins purification and concentration process.
Declaration of interest
This research was funded within of National Health Research Program “Viasan” of the Ministry of Education and Research, Bucharest, Romania.
References
- Theakston RD, Warrell DA, Griffiths E. Report of a WHO workshop on the standardization and control of antivenoms. Toxicon 2003;41:541–57.
- Fry BG, Winkel KD, Wickramaratna JC, et al. Effectiveness of snake antivenom: species and regional venom variation and its clinical impact. J Toxicol 2003;22:23–34.
- Laing GD, Yarleque A, Marcelo A, et al. Preclinical testing of three South American antivenoms against the venoms of five medically-important Peruvian snake venoms. Toxicon 2004;44:103–6.
- Jones RG, Landon J. Enhanced pepsin digestion: a novel process for purifying antibody F(ab')(2) fragments in high yield from serum. J Immunol Methods 2002;263:57–74.
- Jones RG, Landon J. A protocol for 'enhanced pepsin digestion': a step by step method for obtaining pure antibody fragments in high yield from serum. J Immunol Methods 2003;275:239–50.
- Chotwiwatthanakun C, Pratanaphon R, Akesowan S, et al. Production of potent polyvalent antivenom against three elapid venoms using a low dose, low volume, multi-site immunization protocol. Toxicon 2001;39:1487–94.
- Ariaratnam CA, Meyer WP, Perera G, et al. A new monospecific ovine Fab fragment antivenom for treatment of envenoming by the Sri Lankan Russell's viper (Daboia Russelii Russelii): a preliminary dose-finding and pharmacokinetic study. Am J Trop Med Hyg 1999;61:259–65.
- Leon G, Stiles B, Alape A, et al. Comparative study on the ability of IgG and F(ab')2 antivenoms to neutralize lethal and myotoxic effects induced by Micrurus Nigrocinctus (coral snake) venom. Am J Trop Med Hyg 1999;61:266–71.
- Theakston RDG, Reid HA. Development of simple standard assay procedures for the characterization of snake venoms. Bull WHO 1983;61:949–56.
- WHO. Progress in the characterization of venoms and standardization of antivenoms. W.H.O. Offset Publication No. 58. Geneva: WHO; 1981.
- Riviere G, Choumet V, Audebert F, et al. Effect of antivenom on venom pharmacokinetics in experimentally envenomed rabbits: toward an optimization of antivenom therapy. J Pharmacol Exp Ther 1997;281:1–8.
- Riviere G, Choumet V, Saliou B, et al. Absorption and elimination of viper venom after antivenom administration. J Pharmacol Exp Ther 1998;285:490–5.
- de Roodt AR, Dolab JA, Segre L, et al. The immunochemical reactivity and neutralizing capacity of polyvalent Vipera (European) antivenom on enzymatic and toxic activities in the venoms of crotalids from Argentina. J Venom Anim Toxins 1999;5:67–83.
- Peres CM, Bastos MF, Ferreira J, Sartori A. Detection and neutralization of venom by ovine antiserum in experimental envenoming by Bothrops jararaca. J Venom Anim Toxins Incl Trop Dis 2006;12:124–36.
- Maria WS, Cambuy MO, Costa JO, et al. Neutralizing potency of horse antibothropic antivenom. Correlation between in vivo and in vitro methods. Toxicon 1998;36:1433–9.
- Alape-Giron A, Miranda-Arrieta K, Cortes-Bratti X, et al. A comparison of in vitro methods for assessing the potency of therapeutic antisera against the venom of the coral snake Micrurus nigrocinctus. Toxicon 1997;35:573–81.
- Donlon J, Fottrell PF. Quantitative determination of intestinal peptide hydrolase activity using L-amino acid oxidase. Clin Chim Acta 1971;33:345–50.
- Capitanescu A, Capitanescu C, Bisceanu C. Kit of analysis of amino acids using L-amino acid oxidase and peroxidase. Roumanian Biotechnol Lett 2005;10:2179–84.
- Ben Salah R, Zouari N, Reinbolt J, Mejdoub H. Purification of Turkey pancreatic phospholipase A2. Biosci Biotechnol Biochem 2003;67:2139–44.
- Murata Y, Satake M, Suzuki T. Studies on snake venom. Distribution of proteinase activities among Japanese and Formosa snake venoms. J Biol Chem Tokyo 1963;53:431–7.
- Capitanescu C, Ganescu I, Magearu V, Rotariu L. Proteolytic activity of crude dried venom from Vipera ammodytes. Roumanian Biotechnol Lett 2000;5:35–8.