Abstract
Reactive oxygen species (ROS) play an integral role in the pathogenesis of most diseases. This work presents the design and synthesis of novel 2-phenylquinazolin-4-amine derivatives (2–12) and evaluation of their NAD(P)H:quinone oxidoreductase 1 (NQO1) inducer activity in murine cells. Also, molecular docking of all the new compounds was performed to assess their ability to inhibit Keap1–Nrf2 protein–protein interaction through occupying the Keap1–Nrf2-binding domain which biologically leads to a consequent Nrf2 accumulation and enhanced gene expression of NQO1. Docking results showed that all compounds have the ability to interact with Keap1; however compound 7, the most active compound in this study, showed more interactions with key amino acids.
Introduction
Excessive unbalanced production of reactive oxygen species (ROS), namely superoxide anion radical (), singlet oxygen (1O2), hydroxyl radical (•OH) and hydrogen peroxide (H2O2), has been well established to be associated with many pathological conditions including atherosclerosis, stroke, diabetes, Alzheimer’s disease, cancer and inflammationCitation1. In a counter action and to maintain homeostasis, living cells combat the deleterious effects of ROS by an arsenal of biochemical antioxidantsCitation2,Citation3. Under oxidative stress conditions; where the balance between ROS and biochemical antioxidants is lost due to over production of the former or under availability of the latter; ROS can attack and damage cellular membrane lipids, DNA nucleotidesCitation4, proteins’ sulphydryl groupsCitation5 and ribonucleoproteinsCitation6. Among the most important antioxidant mechanisms activated under conditions of oxidative stress is the Kelch-like ECH-associated protein 1 (Keap1)–nuclear factor erythroid 2-related factor 2 (Nrf2)-antioxidant response elements (AREs) pathwayCitation7. This pathway is associated with the expression of more than 100 cytoprotective genes and their respective enzymes, one of which is NAD(P)H:quinone oxidoreductase 1 (NQO1, EC 1.6.99.2). Under homeostatic conditions, Nrf2 is continuously targeted for ubiquitination and proteasomal degradation by Keap1. However, upon increased oxidative load, reactive cysteine residues within Keap1 serve as sensors for oxidants, leading to loss of its ability to target Nrf2 for degradation, subsequent accumulation of the transcription factor, activation of the pathway and increased transcription of cytoprotective antioxidant proteinsCitation8–10.
Medicinal chemists view the Keap1–Nrf2 protein–protein interaction as a critical point of the pathway that can be targeted for intervention and for potential management of a variety of oxidative stress-related ailments like cancer, Parkinson’s, Alzheimer’s and diabetesCitation11–14. Design of non-covalent small molecule modulators of the Keap–Nrf2 interaction has been widely exploredCitation15,Citation16. Quinazoline-based nitrogenous heteroaryls are an excellent reservoir of bioactive substances where a number of biological activities have been associated with antioxidant activityCitation17–19.
In continuation of our quest for new cytoprotective agentsCitation20,Citation21, we herein report the synthesis, NQO1 inducer activity and Keap1 inhibitory in silico screening results for a novel class of N-aryl substituted 4-amino-2-phenylquinazolines.
Materials and methods
Chemistry
Melting points (uncorrected) were determined in open capillary tube on a Gallen Kamp melting point apparatus (Sanyo Gallen Kamp, Leicestershire, UK). Precoated silica gel plates (Kieselgel 0.25 mm, 60 F254, Merck, Darmstadt, Germany) were used for thin layer chromatography. A developing solvent system of chloroform/methanol (8:2) was used and the spots were detected by ultraviolet light. IR spectra (KBr disc) were recorded using an FT-IR spectrophotometer (Perkin Elmer, Waltham, MA). NMR spectra were scanned on a NMR spectrophotometer (Bruker AXS Inc., Geneva, Switzerland), operating at 500 MHz for 1H and 125.76 MHz for 13C. Chemical shifts are expressed in δ-values (ppm) relative to TMS as an internal standard, using DMSO-d6 as a solvent. Elemental analyses were done on a model 2400 CHNSO analyzer (Perkin Elmer, Waltham, MA). All the values were within ±0.4% of the theoretical values. All reagents used were of AR grades. The starting material 4-chloro-2-phenylquinazoline (1) was purchased from Sigma (St. Louis, MO) and was directly used for the preparation of target compounds.
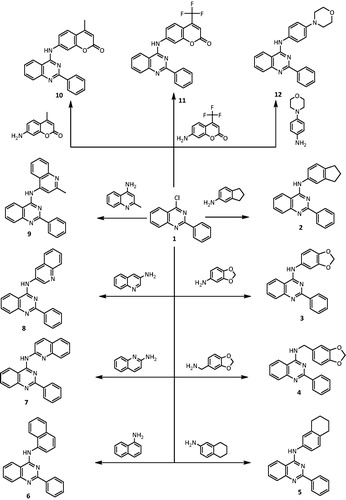
Synthesis of 2-phenyl-4-substituted quinazoline derivatives 2–12
General procedure
A mixture of 2-phenyl 4-chloroquinazoline 1 (2.40 g, 0.01 mol) and different amines (0.01 mol) in dimethylformamide (10 mL) containing a catalytic amount of trimethylamine (three drops) was refluxed for 24 h and then left to cool. The solid product formed was collected by filtration and recrystallized from acetic acid to give the desired quinazoline derivatives 2–12.
N-(2,3-dihydro-1H-inden-5-yl)-2-phenylquinazolin-4-amine (2)
White solid; yield, 76%; m.p. 118–119 °C. IR (KBr, cm−1): 3401 (NH), 3080 (CH arom.), 2976, 2778 (CH aliph.), 1631 (C=N). 1H-NMR (DMSO-d6): ppm 1.9–2.8 [m, 6H, 3CH2, Cyclo], 7.1–9.0 [m, 12H, Ar–H], 11.8 [s, 1H, NH, exchangeable with D2O]. 13C-NMR (DMSO-d6): ppm 25.6, 32.9 (2), 113.1, 121.6, 123.0, 124.4, 125.4, 128.3 (2), 129.3, 129.4, 129.7, 129.8, 133.7 (2), 135.3, 136.1, 142.3, 144.5 (2), 157.3, 159.1. MS m/z (%): 337 (M+) (16.73), 219 (100). Anal. Calcd. for C23H19N3 (337): C, 81.87; H, 5.68; N, 12.45. Found: C, 81.53; H, 5.94; N, 12.14.
N-(benzo[d][1,3]dioxol-5-yl)-2-phenylquinazolin-4-amine (3)
Pale yellow solid; yield, 89%; m.p. 107–108 °C. IR (KBr, cm−1): 3441 (NH), 3081 (CH arom.), 2984, 2874 (CH aliph.), 1617 (C=N). 1H-NMR (DMSO-d6): ppm 6.0 [s, 2H, O–CH2–O], 7.0–8.5 [m, 12H, Ar–H], 9.7 [s, 1H, NH, exchangeable with D2O]. 13C-NMR (DMSO-d6): ppm 101.4, 104.8, 108.6, 114.4, 115.8, 123.3, 126.2 (2), 128.3, 128.5, 128.8 (2), 130.6, 133.5, 133.9, 138.8 (2), 147.4, 150.8, 159.6, 162.7. MS m/z (%): 341 (M+) (3.65), 294 (100). Anal. Calcd. for C21H15N3O2 (341): C, 73.89; H, 4.43; N, 12.31. Found: C, 74.11; H, 4.75; N, 12.65.
N-(benzo[d][1,3]dioxol-5-ylmethyl)-2-phenylquinazolin-4-amine (4)
Tan white solid; yield, 80%; m.p. 180–181 °C. IR (KBr, cm−1): 3238 (NH), 3069 (CH arom.), 2944, 2791 (CH aliph.), 1618 (C=N). 1H-NMR (DMSO-d6): ppm 4.8 [s, 2H, CH2], 5.9 [s, 2H, O–CH2–O], 6.8–8.6 [m, 12H, Ar–H], 10.2 [s, 1H, NH, exchangeable with D2O]. 13C-NMR (DMSO-d6): ppm 44.5, 101.3, 108.4, 108.5, 113.4, 120.9, 124.2, 127.2 (2), 129.1 (2), 132.4 (2), 132.9, 133.3, 134.8 (2), 146.5, 146.7, 147.7, 158.4, 163.4. MS m/z (%): 355 (M+) (22.15), 233 (100). Anal. Calcd. for C22H17N3O2 (355): C, 74.35; H, 4.82; N, 11.82. Found: C, 74.08; H, 4.59; N, 12.10.
2-Phenyl-N-(5,6,7,8-tetrahydronaphthalen-1-yl)quinazolin-4-amine (5)
White solid; yield, 77%; m.p. 192–193 °C. IR (KBr, cm−1): 3412 (NH), 3068 (CH arom.), 2922, 2782 (CH aliph.), 1609 (C=N). 1H-NMR (DMSO-d6): ppm 1.6–2.8 [m, 8H, 4CH2 Cyclo], 7.0–9.0 [m, 12H, Ar–H], 11.4 [s, 1H, NH, exchangeable with D2O]. 13C-NMR (DMSO-d6): ppm 22.6 (2), 25.0, 29.6, 113.0 (2), 125.1, 125.3, 125.8, 128.0, 128.7 (2), 129.1 (2), 129.3 (2), 133.1, 134.1, 135.7, 135.8, 138.4, 157.7, 160.1, 163.4. MS m/z (%): 351 (M+) (7.13), 294 (100). Anal. Calcd. for C24H21N3 (351): C, 82.02; H, 6.02; N, 11.96. Found: C, 82.34; H, 5.81; N, 12.21.
N-(naphthalen-1-yl)-phenylquinazolin-4-amine (6)
White solid; yield, 79%; m.p. 143–145 °C. IR (KBr, cm−1): 3401 (NH), 3100 (CH arom.), 2981, 2780 (CH aliph.), 1629 (C=N). 1H-NMR (DMSO-d6): ppm 7.1–8.9 [m, 16H, Ar–H], 11.2 [s, 1H, NH, exchangeable with D2O]. 13C-NMR (DMSO-d6): ppm 113.7, 119.4, 122.4, 122.8, 124.1, 124.7, 125.2, 126.0, 126.2, 126.5 (2), 127.3, 127.4, 128.2, 128.7, 129.0, 130.0, 131.8, 133.1, 134.1, 135.0, 158.6, 160.8, 163.4. MS m/z (%): 347 (M+) (11.23), 269 (100). Anal. Calcd. for C24H17N3 (347): C, 82.97; H, 4.93; N, 12.10. Found: C, 82.64; H, 5.21; N, 11.86.
2-Phenyl-N-(quinolin-2-yl)quinazolin-4-amine (7)
White solid; yield, 69%; m.p. 191–192 °C. IR (KBr, cm−1): 3411 (NH), 3024 (CH arom.), 2977, 2839, 2780 (CH aliph.), 1635 (C=N). 1H-NMR (DMSO-d6): ppm 7.1–8.9 [m, 15H, Ar–H], 9.1 [s, 1H, NH, exchangeable with D2O]. 13C-NMR (DMSO-d6): ppm 112.6, 116.7, 117.2, 123.1, 126.6 (2), 127.7, 127.8 (2), 128.0 (2), 129.2 (2), 129.3, 129.5, 129.8, 133.1, 135.4, 138.4 (2), 157.1, 161.7, 164.8. MS m/z (%): 348 (M+) (24.07), 209 (100). Anal. Calcd. for C23H16N4 (348): C, 79.29; H, 4.63; N, 16.08. Found: C, 79.55; H, 4.38; N, 16.36.
2-Phenyl-N-(quinolin-3-yl)quinazolin-4-amine (8)
White solid; yield, 91%; m.p. 281–282 °C. IR (KBr, cm−1): 3321 (NH), 3059 (CH arom.), 1624 (C=N). 1H-NMR (DMSO-d6): ppm 7.4–9.3 [m, 15H, Ar–H], 10.3 [s, 1H, NH exchangeable with D2O]. 13C-NMR (DMSO-d6): ppm 114.4, 121.4, 124.8, 125.6, 126.3, 126.5, 127.0 (2), 127.9, 128.1, 128.4, 128.9 (2), 129.1, 130.2, 131.8, 132.2, 133.9, 138.2, 144.7, 149.2, 162.7, 163.2. MS m/z (%): 348 (M+) (39.41), 270 (100). Anal. Calcd. for C23H16N4 (348): C, 79.29; H, 4.63; N, 16.08. Found: C, 79.56; H, 4.32; N, 16.29.
N-(2-methylquinolin-4-yl)-2-phenylquinazolin-4-amine (9)
White solid; yield, 88%; m.p. 236–238 °C. IR (KBr, cm−1): 3433 (NH), 3023 (CH arom.), 2976, 2839, 2779 (CH aliph.), 1609 (C=N). 1H-NMR (DMSO-d6): ppm 2.5 [s, 3H, CH3], 7.5–8.0 [m, 14H, Ar–H], 12.5 [s, 1H, NH exchangeable with D2O]. 13C-NMR (DMSO-d6): ppm 19.8, 102.0, 115.5, 119.7, 121.4, 124.2, 126.1, 126.4 (2), 127.0, 127.9, 128.2, 128.9 (2), 129.7, 131.8, 133.1, 133.9, 149.1, 152.8, 153.2, 158.1, 162.7 (2). MS m/z (%): 362 (M+) (24.09), 347 (100). Anal. Calcd. for C24H18N4 (362): C, 79.54; H, 5.01; N, 15.46. Found: C, 79.22; H, 4.87; N, 15.18.
4-Methyl-7-(2-phenylquinazolin-4-ylamino)-2H-chromen-2-one (10)
Orange solid; yield, 78%; m.p. 279–280 °C. IR (KBr, cm−1): 3356 (NH), 3062 (CH arom.), 2946, 2869, 2765 (CH aliph.), 1699 (C=O), 1618 (C=N). 1H-NMR (DMSO-d6): ppm 2.4 [s, 3H, CH3], 6.2 [s, 1H, CH Chromene], 7.5–8.6 [m, 12H, Ar–H], 10.1 [s, 1H, NH exchangeable with D2O]. 13C-NMR (DMSO-d6): ppm 18.5, 108.1 (2), 112.7, 114.5, 115.3, 123.5, 125.9, 126.7 (2), 128.3, 128.7, 129.0 (2), 130.9, 134.0, 138.6, 143.3, 151.0, 153.6, 154.0, 158.1, 159.3, 160.6. MS m/z (%): 379 (M+) (9.64), 294 (100). Anal. Calcd. for C24H17N3O2 (379): C, 75.97; H, 4.52; N, 11.08. Found: C, 75.61; H, 4.87; N, 10.78.
7-(2-Phenylquinazolin-4-ylamino)-4-(trifluoromethyl)-2H-chromen-2-one (11)
Yellow solid; yield, 72%; m.p. 255–257 °C. IR (KBr, cm−1): 3355 (NH), 3058 (CH arom.), 1697 (C=O), 1617 (C=N). 1H-NMR (DMSO-d6): ppm 6.2 [s, 1H, CH Chromene], 7.2–8.6 [m, 12H, Ar–H], 10.1 [s, 1H, NH exchangeable with D2O]. 13C-NMR (DMSO-d6): ppm 106.3, 108.1, 112.6, 112.9, 114.5, 121.4, 123.5, 124.7, 125.9 (2), 126.5, 127.0, 128.3 (2), 129.0, 130.9, 133.1, 143.5, 149.2, 154.0, 158.1, 160.6, 160.7, 162.6. MS m/z (%): 433 (M+) (1.98), 355 (100). Anal. Calcd. for C24H14F3N3O2 (433): C, 66.51; H, 3.26; N, 9.70. Found: C, 66.21; H, 3.54; N, 9.98.
N-(4-morpholinophenyl)-2-phenylquinazolin-4-amine (12)
White solid; yield, 86%; m.p. 228–229 °C. IR (KBr, cm−1): 3334 (NH), 3059 (CH arom.), 2977, 2891, 2860 (CH aliph.), 1617 (C=N). 1H-NMR (DMSO-d6): ppm 2.8–3.7 [m, 8H, 4CH2 Cyclo], 7.0–8.5 [m, 13H, Ar–H], 9.9 [s, 1H, NH exchangeable with D2O]. 13C-NMR (DMSO-d6): ppm 49.2 (2), 66.6 (2), 114.5 (2), 115.4, 123.4 (2), 126.1, 128.3 (2), 128.5 (2), 128.8 (2), 130.6, 131.6, 133.4 (2), 138.9, 148.0, 159.6, 162.7. MS m/z (%): 382 (M+) (12.23), 295 (100). Anal. Calcd. for C24H22N4O (382): C, 75.37; H, 5.80; N, 14.65. Found: C, 75.04; H, 5.44; N, 14.31.
Biological evaluation
Hepa1c1c7 murine hepatoma cells were maintained in a humidified atmosphere at 37 °C, 5% CO2 in α-MEM supplemented with 10% (v/v) fetal bovine serum. Cells (104 per well) were grown for 24 h in 96-well plates. For evaluation of the potential NQO1 inducer activity of the new 2-phenylquiazolin-4-amines 2–12, the medium was replaced with inducer-containing fresh medium, and the cells were further grown for 48 h. There were eight replicates of each treatment of eight different concentrations of inducers. All compounds were dissolved in DMSO and diluted in the cell culture medium at a ratio of 1:1000 prior to screening experiments. The final solvent concentration in the medium was 0.1% (v/v) in all wells. At the end of the treatment period, cell lysates were prepared in digitonin and the specific activity of NQO1 was determined using menadione as a substrate as previously describedCitation22. The Concentration which Doubles the specific activity of NQO1 (CD value) was used as a measure of inducer potency. Mean values for the eight replicate wells are shown for each data point. The standard deviation for each data point was within 5% of the value.
Molecular modeling study
The 3D molecular models of the newly synthesized phenylquinazolinamine derivatives were built according to the default parameters, with the MOE software suite version 10.2009. Geometry optimization as well as a systematic conformational search was carried out to an RMS gradient of 0.01 Å employing the ConfSearch module implemented in MOE. All computations were performed with the Merck Force Field (MMFF94s). To evaluate our compounds’ ability to access and block the Nrf2-binding site of Keap1, a molecular docking study was performed using the crystallographic structure of the Kelch domain of Keap1 obtained from the Protein Data Bank (PDB ID: 4IQK). The protein target was prepared for docking by addition of the missing hydrogens and calculating the partial charges. Internal validation was performed to the native ligand followed by docking of the compounds where the target protein was kept rigid, while ligands were allowed to rotate to accommodate freely inside the protein cavity. Following multiple separate docking simulations using default parameters, the best conformations were chosen based on the combination of S score data, E conformation and appropriate fitting with the relevant amino acids in the binding pocket.
Results and discussion
Chemistry
The aim of this work was to design and synthesize a novel series of quinazolines bearing biologically active heterocyclic moieties to evaluate and explore their NQO1 inducer activity. Therefore, 4-aminoquinazoline derivatives 2–12 were synthesized starting from 4-chloro-2-phenylquinazoline (1) as depicted in Scheme 1. Compounds 2–12 were obtained in good yield via reaction of 1 with the appropriate amine in dry dimethyl formamide (Scheme 1). All the synthesized compounds were established on basis of their elemental analyses, IR, 1H-NMR, 13C-NMR and mass spectral data. IR spectra of compounds 2–12 revealed the presence of the characteristic bands for the NH group ranging from 3441 to 3238 cm−1, the aromatic CH around 3100–3023 cm−1, the C=N group from 1635 to 1609 cm−1. 1H-NMR spectra of compounds 2–12 exhibited signals at δ 12.5–19.1 ppm attributed to the NH group. Further details of the spectroscopic analysis findings of all the synthesized compounds are thoroughly described in the chemistry experimental section.
Biological evaluation
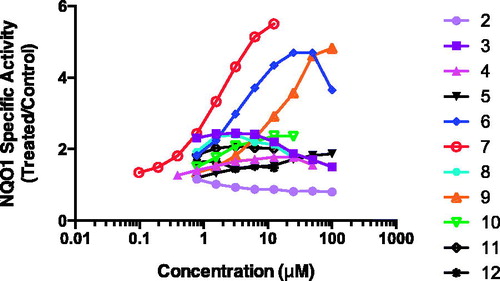
NQO1 inducer activity assessment represented in and revealed that the dihydroindenyl derivative 2, the dioxo bioisostere N-benzo[d][1,3]dioxol-5-yl derivative 3, its methylene spacer-containing derivative 4, the tetrahydronaphthyl derivative 5 and the morpholinophenyl compound 12 were all essentially inactive. In contrast, the naphth-1-yl derivative 6 showed a moderate dose-dependent inducer activity (CD = 1 μM). The 2-methyl-quinolin-4-yl analog 9 displayed a four times weaker inducer potency (CD = 4 μM), indicating that these structural modifications were not in favor of activity as compared to the naphthyl bioisostere 6. The bioisosteres 7 and 8 demonstrated a large difference in activity. Whereas, the quinolin-2-yl derivative 7 had the highest potency among the compounds in this study (CD = 0.4 μM), the activity of the three positional isomer 8 was lower (CD = 0.9 μM); furthermore, the absence of dose-dependence for compound 8 is indicative of low specificity and potential off-target effects. Finally, with regards to the bioisosteric pair 10 and 11 (CD = 3 and 1.6 μM, respectively) it was evident that replacement of the methyl group with a triflouromethyl almost doubled the potency and improved the dose responsiveness. Overall, compound 7 was the most potent inducer (CD = 0.4 μM), and importantly, had the most favorable inducer properties within this series.
Table 1. NQO1 inducer activity and CD values of the test compounds.
Molecular modeling study
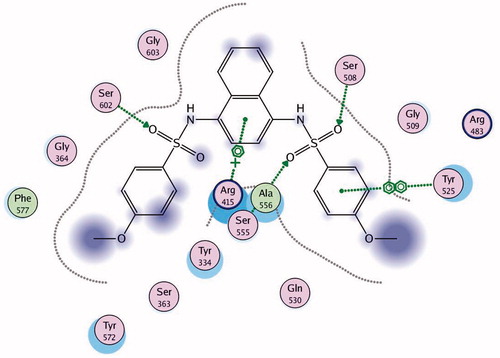
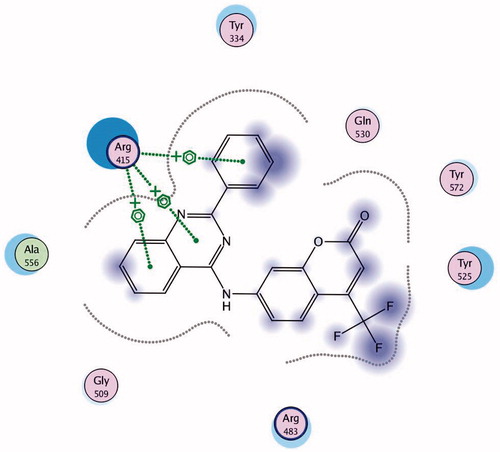
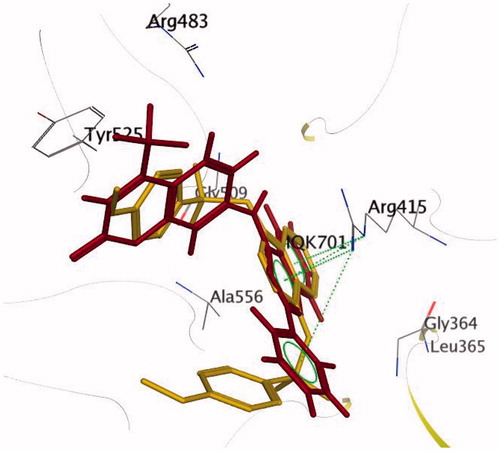
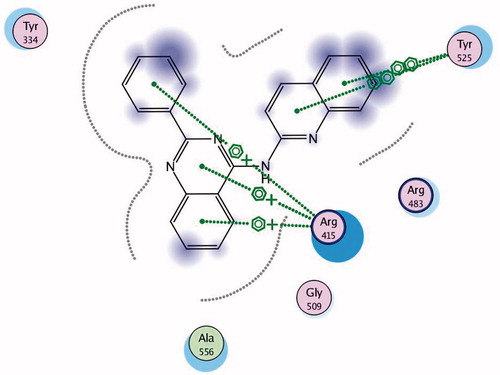
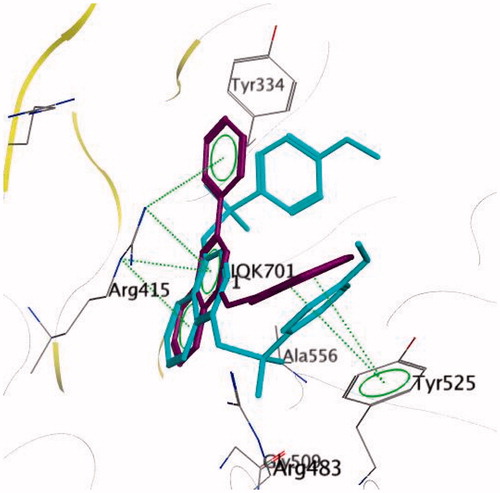
Keap1 is known to bind to Nrf2 promoting its degradation, resulting in low levels of cytoprotective gene products. Various small molecule compounds were reported to bind to the Keap1 Kelch domain and antagonize its activityCitation23–25. For assessing the newly synthesized compounds’ ability to access and block the Kelch domain of Keap1, a molecular docking study was performed using the Kelch domain of Keap1 crystal coordinates obtained from the PDB ID: 4IQK. The main interactions observed between the validated native ligand and the protein target were found to be arene–cation interaction with Arg415, arene–arene interaction with Tyr525 and three hydrogen bonds with Ser602, Ser508 and Ser555 with S= −13.306 kcal/mol and rmsd 0.6635 kcal/mol/Å (). Upon docking of the synthesized compounds, they all showed an arene–cation binding interaction with Arg415 via their aromatic rings. Compound 11 (S = −10.913 kcal/mol) has adopted a conformation allowing the presence of three arene–cation interactions with Arg415 and multiple rings of the ligand ( and ).
Finally, compound 7 that showed the most potent NQO1 inducer activity (CD = 0.4 μM) was the only compound that showed three arene–cation interactions with Arg415 and two arene–arene interactions with Tyr525 indicating a possible correlation between those multiple interactions and the noted high potency (S= −10.7652 kcal/mol) ( and ).
One limitation of our study is the lack of a well-established biological assay for assessing the ability of small molecules to disrupt the Keap1–Nrf2 protein–protein interactions. Therefore, although the molecular docking study suggests that these compounds can potentially bind to Keap1 and disrupt its interaction with Nrf2, further experimental evidence is required to confirm this possibility. Regardless of the precise mode of action in activating Nrf2, the inducer potency of compound 7 in upregulating NQO1 strongly suggests that this compound could be used as a lead for further optimization of the quinazoline scaffold.
Conclusion
In conclusion, this study presents the synthesis of N-substituted 2-phenylquinazolin-4-amine based derivatives 2–12 as cytoprotective NQO1 inducers. The most potent inducer in this study was the analogue 7 which caused a dose-dependent induction of NQO1 with a CD value of 0.4 μM. In silico evaluation of these derivatives as Keap1–Nrf2 protein–protein interaction inhibitors was performed using molecular docking techniques employing MOE software version 10.2009. Derivatives were found to bind to key amino acids in the binding site with a network or arene–arene and arene–cationic interactions. These molecules represent promising quinazoline scaffold-based leads for further optimization as cytoprotective agents and for future investigations into other plausible modes of action.
Declaration of interest
The authors would like to extend their sincere appreciation to the Deanship of Scientific Research at King Saud University for funding of this research through the Research Group Project no. RGP-VPP-302. Maureen Higgins and Albena T. Dinkova-Kostova are also grateful to Cancer Research UK (C20953/A10270 and C20953/A18644) for their financial support.
References
- Waris G, Ahsan H. Reactive oxygen species: role in the development of cancer and various chronic conditions. J Carcinog 2006;5:14
- Scandalios JG. The rise of ROS. Trends Biochem Sci 2002;27:483–6
- Klein JA, Ackerman SL. Oxidative stress, cell cycle, and neurodegeneration. J Clin Invest 2003;111:785–93
- Ahsan H, Ali A, Ali R. Oxygen free radicals and systemic autoimmunity. Clin Exp Immunol 2003;131:398–404
- Knight JA. Diseases related to oxygen-derived free radicals. Ann Clin Lab Sci 1995;25:111–21
- Waris G, Alam K. Attenuated antigenicity of ribonucleoproteins modified by reactive oxygen species. Biochem Mol Biol Int 1998;45:33–45
- Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1–Nrf2–ARE pathway. Annu Rev Pharmacol Toxicol 2007;47:89–116
- Kobayashi M, Li L, Iwamoto N, et al. The antioxidant defense system Keap1–Nrf2 comprises a multiple sensing mechanism for responding to a wide range of chemical compounds. Mol Cell Biol 2009;29:493–502
- McMahon M, Lamont DJ, Beattie KA, Hayes JD. Keap1 perceives stress via three sensors for the endogenous signaling molecules nitric oxide, zinc, and alkenals. Proc Natl Acad Sci USA 2010;107:18838–43
- Dinkova-Kostova AT, Holtzclaw WD, Cole RN, et al. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci USA 2002;99:11908–13
- Li J, Ichikawa T, Janicki JS, Cui T. Targeting the Nrf2 pathway against cardiovascular disease. Expert Opin Ther Targets 2009;13:785–94
- Zhao J, Redell JB, Moore AN, Dash PK. A novel strategy to activate cytoprotective genes in the injured brain. Biochem Biophys Res Commun 2011;407:501–6
- Steel R, Cowan J, Payerne E, et al. Anti-inflammatory effect of a cell-penetrating peptide targeting the Nrf2/Keap1 interaction. ACS Med Chem Lett 2012;3:407–10
- Williamson TP, Amirahmadi S, Joshi G, et al. Discovery of potent, novel Nrf2 inducers via quantum modeling, virtual screening, and in vitro experimental validation. Chem Biol Drug Des 2012;80:810–20
- Hu L, Magesh S, Chen L, et al. Discovery of a small-molecule inhibitor and cellular probe of Keap1–Nrf2 protein–protein interaction. Bioorg Med Chem Lett 2013;23:3039–43
- Zhuang C, Miao Z, Sheng C, Zhang W. Updated research and applications of small molecule inhibitors of Keap1–Nrf2 protein–protein interaction: a review. Curr Med Chem 2014;21:1861–70
- Nesterova NA, Kovalenko SI, Belenichev IF, et al. Formation of combinational library of quinazoline-4-yl-hydrazones with antioxidant activity. Ukraine Med Khim 2004;6:14–21
- Nesterova NA, Kovalenko SI, Karpenkos OV, Belenichev IF. Synthesis and antioxidant activity of 4-arylidenehydrazinoquinazolines. Ukr Farmatsevtichnii Zhurnal (Kiev) 2004;22:5–10
- Al-Omar MA, Al-Rashood ST, El-Subbagh HI, Abdel Hamide SG. Interaction of 2-thio-4-oxo-quinazoline derivatives with guinea pig liver molybdenum hydroxylases, xanthine oxidase and aldehyde oxidase. J Biol Sci 2005;5:370–8
- Ghorab MM, Higgins M, Alsaid MS, et al. Synthesis, molecular modeling and NAD(P)H:quinone oxidoreductase 1 inducer activity of novel cyanoenone and enone benzenesulfonamides. J Enzyme Inhib Med Chem 2014;29:840–5
- Ghorab MM, Higgins M, Dinkova-Kostova AT, et al. Novel thioureido derivatives carrying thione and sulfonamide moieties induce the cytoprotective enzyme NAD(P)H:quinone oxidoreductase 1. Asian J Chem 2014;26:8501–4
- Prochaska HJ, Santamaria AB. Direct measurement of NAD(P)H:quinone reductase from cells cultured in microtiter wells: a screening assay for anticarcinogenic enzyme inducers. Anal Biochem 1988;169:328–36
- Marcotte D, Zeng W, Hus JC, et al. Small molecules inhibit the interaction of Nrf2 and the Keap1 Kelch domain through a noncovalent mechanism. Bioorg Med Chem 2013;21:4011–19
- Magesh S, Chen Y, Hu L. Small molecule modulators of Keap1–Nrf2–ARE pathway as potential preventive and therapeutic agents. Med Res Rev 2012;32:687–726
- Bertrand HC, Schaap M, Baird L, et al. Design, synthesis, and evaluation of triazole derivatives that induce Nrf2 dependent gene products and inhibit the Keap1–Nrf2 protein–protein interaction. J Med Chem 2015;58:7186–94