Abstract
The CSN complex plays a key role in various cellular pathways: through a metalloprotease activity of its Csn5 deneddylating enzyme, it regulates the activity of Cullin-RING ligases (CRLs). Indeed, Csn5 has been found amplified in many tumors, but, due to its pleiotropic effects, it is difficult to dissect its function and the involvement in cancer progression. Moreover, while growing evidences point to the neddylation function as a good target for drug development; specific inhibitors have not yet been developed for the CSN. Here, we propose the yeast Saccharomyces cerevisiae as a model system to screen libraries of small molecules as inhibitors of cullins deneddylation, taking advantage of the unique feature of this organism to survive without a functional CSN5 gene and to accumulate a fully neddylated cullin substrate. By combining molecular modeling and simple genetic tools, we were able to identify two small molecular fragments as selective inhibitors of Csn5 deneddylation function.
Introduction
The COP9 signalosome (CSN) is evolutionary conserved among all eukaryotes, multi-subunit complex with a canonical composition of eight subunits (Csn1–8) found in all multicellular organisms. The CSN regulates the activity of the largest family of ubiquitin E3 ligases, called CRLs (Cullin-RING ubiquitin ligases). Regulation of CRLs by the CSN involves the catalytic activity of NEDD8 hydrolysis from the cullin scaffold subunit of CRLs through a metalloprotease MPN+/JAMM motif within the catalytic subunit, Csn5.
Through a complicated regulation sequence of neddylation and deneddylation, CRLs commit to ubiquitinate their substrates, thus controlling various cellular functions, including cell cycle progression, DNA repair and cell survival.
The yeast Saccharomyces cerevisiae CSNCitation1,Citation2 includes only six subunits: a most conserved catalytic subunit Csn5/Rri1 and five divergent orthologues of canonical subunitsCitation3. Yet, the yeast CSN harbors a canonical cullin-deneddylation activity and is able to deneddylate mammalian cullinsCitation2. Vice versa, mammalian CSN is able to deneddylate yeast cullinsCitation4,Citation5. However, the knockdown of the yeast CSN subunits neither affect viability nor the G1-S transition promoted by the SCF, and it has a modest effect on the turnover of SCF substratesCitation6–8.
The key role of CSN in regulating cullin-RING ligases places it as a central mediator of cellular functions during cancer progression. In particular, the overexpression of CSN5 gene has been found in many tumors, including breast cancersCitation9,Citation10, and growing evidences point to a fundamental role of Csn5 as a key driver of epithelial transformationCitation11. Csn5 could exert its positive function in cancer progression in different ways: first, as part of the CSN, Csn5 is an important activator of MYC activityCitation12; second, in a small complex or a free form, Csn5 induces the cytoplasmic export of p27 (a tumor suppressor gene) that is causing its ubiquitination and proteasomal degradationCitation13–15; third, in monomeric form, Csn5 binds and modulates signaling proteins and transcription factors, such as hypoxia-inducible factor-1αCitation16. This landscape makes difficult to dissect/identify Csn5 role in specific tumors due to its pleiotropic function.
In the era of pharmacogenomic and personalized medicine, the targeting of specific proteins with designed molecules became possible. Yet, up-to-date, no specific inhibitors were selected for Csn5, even if the CSN pathway has gained increasing attention as a target for anticancer moleculesCitation17,Citation18. Indeed, the Csn5 study is challenging due to mammalian embryonic lethality of the CSN5 deletion and the instability of the protein if overespressedCitation19. Recently, the deneddylating activity of the CSN has been described as a novel target of doxycyclineCitation20.
Here, we propose the yeast Saccharomyces cerevisiae as a model system to select small molecules to target the deneddylating function of Csn5.
Methods
Yeast strains
Saccharomyces cerevisiae strains W303 (MATa leu2–3,112, trp1–1, can1–100, ura3–1, ade2–1, his3–11,15), Δcsn5 (MATa leu2–3,112, trp1–1, can1–100, ura3–1, ade2–1, his3–11,15, csn5Δ::KanMX), Δrub1 (MATa leu2–3,112 trp1–1, can1–100, ura3–1, ade2–1, his3–11,15, rub1Δ::KanMX) and rpn11-m1 (MATa leu2–3,112, trp1–1, can1–100, ura3–1, ade2–1, his3–11,15, rpn11-m1) were used in this work. YPD (1% bacto peptone, 1% yeast extract and 2% glucose) was used as yeast culture-rich medium.
Cells treatments
Cells grown in rich media were diluted to 1 × 107 cells/ml and 200 μl aliquots were placed in 96-wells microplate. Each aliquot was treated with different concentrations of the indicated compounds and then incubated shaking at 28 °C. After 20 h OD600 for each well was measured and plotted.
Western blot
Ten milliliter of cells grown to 1 × 107 cell/ml were harvested by centrifugation, re-suspended in 1 ml of YPD containing 2 mM of the indicated compounds or 2% dimethyl sulfoxide (DMSO), and then incubated shaking for 2 h at 28 °C. Extracts were prepared from this cells following trichloroacedic acid (TCA) protein precipitation.
Proteins were resolved on 10% Tris–glycine PAGE,Citation34 transferred to PVDF membranes and immunoblotted using the α-CDC53 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) or α-ubiquitin (Sigma-Aldrich, St. Louis, MO). The secondary antibodies were anti-Goat (Santa Cruz Biotechnology and anti-Rabbit (Santa Cruz Biotechnology). The proteins were visualized utilizing the Bio-Rad’s ChemiDoc XRS + system, according to manufacturer’s instructions.
RNA extraction
RNA was extracted from cell cultures at OD600= 1 (strains were grown in YPD medium at 28 °C). Cells were suspended in 1 mL of AE buffer (50 mM sodium acetate pH 5, 10 mM EDTA) (AppliChem GmbH, Darmstadt, Germany), centrifuged and suspended in 0.4 mL of AE buffer plus 1% (w/v) SDS (AppliChem GmbH). Cells were lysed with phenol:chloroform (5: 1, pH 4.7, Sigma-Aldrich), heated at 65 °C for 10 min, transferred at −80 °C for 10 min and the aqueous phase was separated by centrifugation. After a second extraction with phenol:chloroform (24: 1, pH 5.2, Sigma-Aldrich), RNA was precipitated with ethanol, dried and suspended in sterile water. RNA quantity and purity were assessed with a Nanodrop ND-1000 spectrophotometer (Thermo Scientific, Waltham, MA) at 260 nm and at 260/230, 260/280 nm ratios, respectively. RNA integrity was assessed by electrophoresis on ethidium bromide stained 1% agarose-formaldehyde gels.
RT real-time PCR
1 mg of total RNA extracted from cell cultures, 1 μg RNA was reverse transcribed using 200 ng of 16mers oligo dT (Life Technologies, Carlsbad, CA) with SuperScript III First-Strand Synthesis System for RT-PCR (Life Technologies), according to the manufacturer’s instructions. cDNA served as template for subsequent real-time PCR reactions that were set up in duplicate for each sample using the SensiMix SYBR Mix (Bioline, London, UK) and an Applied Biosystems Prism 7300 Sequence Detector. The reaction mixtures were kept at 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. The threshold cycle (Ct) was calculated using the Sequence Detector Systems version 1.2.2 (Life Technologies) by determining the cycle number at which the change in the fluorescence of the reporter dye (DRn) crossed the threshold. To synchronize each experiment, the baseline was set automatically by the software. Relative quantification was carried out with the 2−ddct method, using the abundance of actin transcript as endogenous housekeeping control. Data were statistically analyzed by Student’s t-test.
Molecular modeling directions
Molecular dynamics simulation
Crystal structure of Csn5/Jab1 enzyme (PDB code: 4F7O) was downloaded from the Protein Data Bank (www.rcsb.org/pdb)Citation21. The chain A from the Csn51–257 crystal structure was used as the starting structure, after manual replacement of Arg106 with Ala (Csn5_R106A mutant). The system was solvated by means of the System Builder tool of DesmondCitation22,Citation23 with a periodic orthorhombic box of explicit TIP3P water molecules. Charge was neutralized by adding three Cl− ions. The entire system of 35041 atoms was relaxed by energy minimization with 2000 maximum iteration steps and a convergence threshold of 1.0 kcal/mol/Å. The minimization procedure is performed with a steepest-descent algorithm (SD) until a gradient threshold of 25 kcal/mol/Å is reached, then the LBFGS algorithm is used. The OPLS2005 force field was used. A weak harmonic restraint of 20 kcal/mol/Å2 was applied to the zinc atom. Molecular dynamics simulation was performed at constant T and P (i.e. NTP simulation), with the OPLS 2005 force field and the following parameters: simulation time = 40 ns, temperature = 300 K, pressure = 1.013 bar. A weak harmonic restraint of 20 kcal/mol/Å2 was applied to both the zinc atom and the atoms directly involved in zinc coordination, namely the two nitrogen atoms of His138 and His140 and the two oxygen atom of Asp151. Other parameters were kept to their default value. The distance between the zinc atom and Ala106 was monitored by the simulation event analysis tool of Desmond.
Molecular docking
The receptor was prepared with the MAKE_RECEPTOR graphical utility implemented in the OEDocking suiteCitation24 of OpenEye, starting from the selected frame of previous molecular dynamics simulation. A box volume of 5473 Å3 with dimensions 17.33 × 16.33 × 19.33 Å covering the accessible surface of the Csn5 catalytic groove was used. A constraint on the zinc atom was also added. Molecules used in the docking procedure were extracted from an in house library, by choosing small fragments endowed with a molecular weight up to 300 Da. Selected structures were further filtered with the FILTER application of OpenEyeCitation25 for the presence of the carboxylate moiety and the protonation state of each molecule was assigned by using the FIXPKA application of QUACPAC of OpenEyeCitation26. Conformational analysis of the database was carried out with the OMEGA software of OpenEye (Santa Fe, NM)Citation27,Citation28, by keeping a maximum of 500 conformations for each molecule. Molecular docking was performed by means of the FRED software (Santa Fe, NM) implemented in the OEDocking suite of OpenEyeCitation29, by saving the best five poses for each molecule and only for the first 50 best scored molecules. Other parameters were kept to their default value.
Energy minimization
Docking-selected complexes were further relaxed in a box of TIP3P explicit water molecules by energy minimization with Desmond. The OPLS2005 force field was used. A weak harmonic restraint of 20 kcal/mol/Å2 was applied to both the zinc atom and the atoms directly involved in zinc coordination, namely the two nitrogen atoms of His138 and His140 and the two oxygen atom of Asp151. Energy minimization was performed with 2000 maximum iteration steps and a convergence threshold of 1.0 kcal/mol/Å. The minimization procedure is performed with a SD until a gradient threshold of 25 kcal/mol/Å is reached, then the LBFGS algorithm is used.
pka and atom charge calculations
pka values were determined by the software Marvin Sketch of ChemAxon (Budapest, Hungary)Citation30. Atom charges were calculated by the Antechamber/Mopac software implemented in Amber12 (San Francisco, CA)Citation31.
Results and discussion
The MPN+/JAMM domain of Csn5 are conserved in evolution across all eukaryotic phyla, including S. cerevisiaeCitation32,Citation33. CRLs in evolution and it is the metalloprotease domain responsible for the cleavage of Nedd8 from Cullin1 (Cdc53 in S. cerevisiae), the scaffold protein of CRL1 (a.k.a SCF)Citation4,Citation34. Taking advantage of the crystal structure of the MPN+/JAMM motif and of the human Csn5, namely Csn5/Jab1 (PDB code: 4F7O)Citation21,35–36, we established a structure-based computational strategy to discover small molecular fragments as potential inhibitors of the enzyme catalytic function (the computational screening workflow is described in Supplementary Figure S1). First, analysis of the above mentioned crystallographic structure revealed that the molecular segment named Ins-1 obstructs the accessibility to the Zn-coordinated active site of Csn5 by means of a salt-bridge between Asp151 and the Ins-1 residue Arg106. Notably, Arg106 replacement with threonine, alanine or glycine, restores the catalytic activityCitation21. Therefore, to overcome this structural obstacle to the design of Csn5 inhibitors, and following the procedure already described by Echalier et al. (2013)Citation21, we mutated Arg106 to alanine in silico in the crystallographic structure of Csn5 (Supplementary Figure S2A). The such generated Csn5_R106A mutant was furthermore relaxed by energy minimization and subsequently studied by means of molecular dynamics (MD) simulations, which showed the release of the Ins-1 segment from the Zn-coordinated active site during MD trajectories, in agreement with previous findingsCitation21. The distance between the catalytic Zn(II) ion and the Ins-1 residue Ala106, was monitored during MD simulations, reaching the maximum values of 20.7 Å that corresponds to a noticeable accessibility of the active site (Supplementary Figure S2B). Overall, the R106A mutation in silico of Csn5 followed by MD simulations provided a conformation of the enzyme that resembles the catalytically competent metalloprotease. The frame corresponding to the maximum distance between Zn(II) and Arg106 was used as rigid receptor in the subsequent structure-based virtual screening (Supplementary Figure S2B).
To this end, an in house library of natural compounds and their derivatives was used. It is worth mentioning that the library is endowed with a significant chemical diversity and has been already employed with success in previous hits and leads discovery studiesCitation37,Citation38. Based on limited structural data on the Csn5 enzyme, the lack of reference inhibitors, and the presence of a Zn(II) ion within the catalytic site, the library was filtered to select only small molecular fragments, in accordance with the “Rule of Three”Citation39, which are endowed with a well-known zinc-binding groupCitation40. The selected subset of fragments (Supplementary Figure S3) was then virtually screened against the MD-refined structure of the Csn5 enzyme by means of molecular docking with FRED from OpenEyeCitation41–43. Docking complexes were further relaxed by energy minimization. Coupling visual inspection with the ranking according with the Chemgauss4 score provided 11 small molecular fragments (Supplementary Figure S4) as potential inhibitors of Csn5 enzyme, which were submitted to functional investigations.
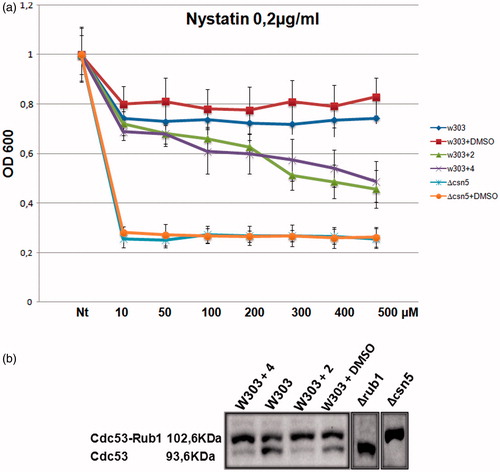
We reasoned that we could take advantage of the versatility of the yeast cells to screen for Csn5 inhibitors and for this purpose we first searched for a rapid test to mimic/verify Csn5 inhibition. In S. cerevisiae, the Δcsn5 strain is viable allowing us to test the phenotypes caused by the CSN5 deletion in presence of a variety of compounds (Supplementary Figure S5). Between the 10 tested compounds, only nystatin produced a clear phenotype in the deleted csn5 strain, compared with the wild-type yeast strain (W303), suitable for inhibitors screening. Nystatin binds to ergosterol, a major component of the fungal cell membrane forming pores and causing yeast cells death; the sensitivity phenotype of Δcsn5 is a consequence of its altered lipid metabolism and indeed we previously demonstrated, by transcriptomic and proteomic experiments, that the Csn5 protein is involved in the modulation of genes controlling the aminoacid and lipid metabolism in yeastCitation34. We reasoned that if the 11 molecules selected by molecular docking inhibit the Csn5 function, we should see a drug sensitive phenotype of the wild-type cells in presence of the selected molecules plus 0.2 μg/ml nystatin (Supplementary Figure S6). Indeed, all the 11 compounds produced a range of sensitivity to nystatin in the wild-type cells from compound 9 (the less sensitive) to compound 4 (the most sensitive), indicating that the molecules could be effective in interfering with the deneddylation function. To test this hypothesis, we performed a western blot analysis to assess the deneddylation activity on Cdc53, which is a major target of Csn5, in a W303 strain treated with the 11 molecules in YPD medium (Supplementary Figure S7). The results showed that molecules 2 and 4 enhanced the sensitivity to nystatin () and inhibited the deneddylation activity of Csn5 at similar concentration (). We refer to compound 2 and 4 as Csn5 specific inhibitors from now on.
Figure 1A. Dose-response curve obtained with molecules 2 and 4. W303 wild-type strain and Δcsn5 were grown in YPD medium and W303 treated over night with molecules 2 and 4 or with DMSO only (at the same concentration used to dissolve the compounds). Nt indicates non-treated with nystatin. The compounds 2 and 4 enhance sensitivity to nystatin mimicking the Δcsn5 behavior. The drastic sensitivity of Δcsn5 cells is due to the complete absence of Csn5 enzyme in these cells.
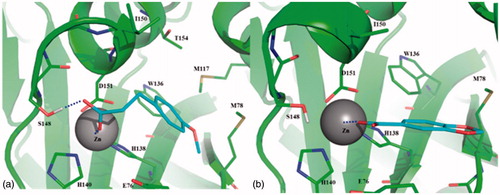
According with molecular modeling, molecules 2 and 4 were predicted to bind and coordinate the catalytic Zn(II) ion by the carboxylic group. In 2, this moiety also establishes a H-bond interaction with the side chain of Ser148 () that, together with Glu76, is known to coordinate a catalytic water molecule in the inactive form of the enzyme. Interestingly, the geometry of coordination to the catalytic Zn(II) ion of 2 and 4 is very common in crystallographic structures of ligand-bound metalloproteases (just as e.g. see PDBs: 1ZZ0, 2WO8, 1OS2)Citation45–48. The aromatic portion of 2 was found to interact in a hydrophobic cleft bounded by Trp136, Met117, Met78, Thr154 and Ile150. The same hydrophobic interactions can be assessed for 4, although to a lesser extent (). Finally, as 10 out of 11 tested fragments bear a carboxylic functional group, it was found to be crucial for Zn-coordination, thus becoming a key structural feature of Csn5 inhibitors. Moreover, according with literature data, in case of metal coordination the higher the pKa values of the carboxylic group (i.e. the lower the acidity), the better are donor properties of the carboxylate ion due to higher electron density on the oxygen atomsCitation49. Noteworthy, pKa of 2 and 4 resulted higher or equal to that calculated for other fragments (Supplementary Information Table S1), thus suggesting that pKa coupled with structural features described above and reported in may be exploited for optimizing Csn5 inhibitors.
Figure 2. Binding mode of 2 (a) and 4 (b) predicted by molecular docking and energy minimization. Csn5 is shown as cartoon. Residues within 4 Å from the ligand are labeled and showed as thin sticks. Zn(II) ion is labeled and showed as sphere. Ligand is shown as thin sticks. Metal coordination of 2 and 4, as well as the H-bond between the carboxylate of 2 and the side chain of Ser148 are reported in dashed line.
The Csn5 has an ortholog, the proteasomal deubiquitinating enzyme Rpn11. These two proteins share a similar MPN+/JAMM domain, so, even if the molecules were specifically designed for Csn5, we cannot exclude that in vivo the molecules could affect the Rpn11 function. In order to verify that the deneddylating inhibition is a direct effect of the molecules in the Csn5 catalytic site and it is not due to a block of Rpn11 proteasomal function, we looked at the profile of ubiquitinated proteins in the cells treated with the inhibitor 2; as a control we used the proteasomal rpn11-m1 mutant, which accumulates polyubiquitinated proteins due to the block of degradationCitation50; the result showed that cells treated with inhibitor no. 2 did not accumulate polyubiquitinated proteins after 30 or 60 min of treatment, indicating that the inhibition was specific for cullin deneddylation function (Supplementary Figure S8).
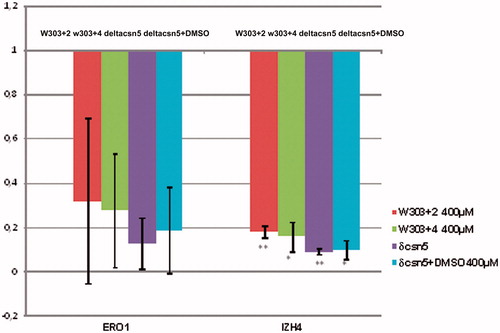
The transcriptomic profile of Δcsn5 strain has been previously definedCitation34. In order to assess if the catalytic inhibition of Csn5 is able to recapitulate some of the transcriptional effects of Csn5 deletion, we quantified by real-time RT-PCR the expression of two genes (ERO1 and IZH2), which are typically down-regulated in Δcsn5 strains, in the W303 strain treated with inhibitors 2 and 4. Indeed we observed that in the presence of these compounds, the mRNA of both genes is reduced in an extent very similar to that observed in the isogenic Δcsn5 strain (). This result suggests that the transcriptional effects observed in the deleted strain are due to the loss of Csn5 catalytic activity.
Figure 3. Real-time RT-PCR analysis of ERO1 and IZH4 mRNA levels after 2 h of treatment with 400 μM inhibitors 2 or 4. The mRNA levels were normalized to those observed in the WT strain grown in presence of DMSO. The mRNA level in the Δcsn5 strain is shown for comparison. The values represent the average of two independent experiments, standard deviation is indicated. Asterisks indicate that in the case of IZH4 the changes are statistically significant according to Student’s t-test results (*p < 0.05; **p < 0.01). In the case of ERO1, although the same tendency is evident, data show a higher variability.
The study of Csn5 function is greatly complicated by its multiple roles as single protein or in complex with CSN partners, some of which do not require its catalytic function. The availability of specific functional inhibitors is therefore essential to identify the effects directly associated with the complex-dependent cullin-deneddylation activity.
Supplementary material available online
IENZ_1160901_supplementary_material_Cirigliano.pdf
Download PDF (506.3 KB)Acknowledgements
We thank Giovanna Serino for helpful scientific discussion. The authors wish also to thank the OpenEye Free Academic Licensing Program for providing a free academic license for molecular modeling and chemoinformatics software.
Declaration of interest
The authors report no declarations of interest. A.C. was supported by Associazione Gian Franco Lupo “Un sorriso alla vita” Onlus. This work is supported by Israel Ministry of Science and Technology (MOST) – Italy Ministry of Foreign Affairs (MAE) grant 3–9022 (to R.N., T.R. and E.P.) and by Ateneo 2014 - prot. C26A14XZC8.
References
- Yu Z, Kleifeld O, Lande-Atir A, et al. Dual function of Rpn5 in two PCI complexes, the 26S proteasome and COP9 signalosome. Mol Biol Cell 2011;22:911–20
- Pick E, Golan A, Zimbler JZ, et al. The minimal deneddylase core of the COP9 signalosome excludes the Csn6 MPN-domain. PLoS One 2012;7:e43980
- Pick E, Hofmann K, Glickman MH. PCI complexes: beyond the proteasome, CSN, and eIF3 Troika. Mol Cell 2009;35:260–4
- Cope GA, Suh GS, Aravind L, et al. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 2002;298:608–11
- Wee S, Hetfeld B, Dubiel W, Wolf DA. Conservation of the COP9/signalosome in budding yeast. BMC Genet 2002;3:15
- Rabut G, Le Dez G, Verma R, et al. The TFIIH subunit Tfb3 regulates cullin neddylation. Mol Cell 2011;43:488–95
- Maytal‐Kivity V, Piran R, et al. COP9 signalosome components play a role in the mating pheromone response of S. cerevisiae. EMBO Reports 2002;3:1215–21
- Zemla A, Thomas Y, Kedziora S, et al. CSN- and CAND1-dependent remodelling of the budding yeast SCF complex. Nat Commun 2013;4:1641
- Richardson KS, Zundel W. The emerging role of the COP9 signalosome in cancer. Mol Cancer Res 2005;3:645–53
- Shackleford TJ, Claret FX. JAB1/CSN5: a new player in cell cycle control and cancer. Cell Division 2010;5:26
- Adler AS, Littlepage LE, Lin M, et al. CSN5 isopeptidase activity links COP9 signalosome activation to breast cancer progression. Cancer Res 2008;68:506–15
- Adler AS, Lin M, Horlings H, et al. Genetic regulators of large-scale transcriptional signatures in cancer. Nat Genet 2006;38:421–30
- Tomoda K, Kubota Y, Kato JY. Degradation of the cyclin-dependent-kinase inhibitor p27Kip1 is instigated by Jab1. Nature 1999;398:160–5
- Pan Y, Zhang Q, Tian L, et al. Jab1/CSN5 negatively regulates p27 and plays a role in the pathogenesis of nasopharyngeal carcinoma. Cancer Res 2012;72:1890–900
- Pick E, Bramasole L. Moonlighting and pleiotropy within two regulators of the degradation machinery: the proteasome lid and the CSN. Biochem Soc Trans 2014;42:1786–91
- Bae MK, Ahn MY, Jeong JW, et al. Jab1 interacts directly with HIF-1alpha and regulates its stability. J Biol Chem 2002;277:9–12
- Luo Z, Yu G, Lee HW, et al. The Nedd8-activating enzyme inhibitor MLN4924 induces autophagy and apoptosis to suppress liver cancer cell growth. Cancer Res 2012;72:3360–71
- Abidi N, Xirodimas DP. Regulation of cancer-related pathways by protein NEDDylation and strategies for the use of NEDD8 inhibitors in the clinic. Endocr-Relat Cancer 2015;22:T55–70
- Mori M, Yoneda-Kato N, Yoshida A, Kato JY. Stable form of JAB1 enhances proliferation and maintenance of hematopoietic progenitors. J Biol Chem 2008;283:29011–21
- Pulvino M, Chen L, Oleksyn D, et al. Inhibition of COP9-signalosome (CSN) deneddylating activity and tumor growth of diffuse large B-cell lymphomas by doxycycline. Oncotarget 2015;6:14796
- Cope GA, Deshaies RJ. COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell 2003;114:663–71
- Lyapina S, Cope G, Shevchenko A, et al. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science 2001;292:1382–5
- Ambroggio XL, Rees DC, Deshaies R. J. JAMM: a metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol 2004;2:E2
- Echalier A, Pan Y, Birol M, et al. Insights into the regulation of the human COP9 signalosome catalytic subunit, CSN5/Jab1. Proc Natl Acad Sci USA 2013;110:1273–8
- Tran HJ, Allen MD, Löwe J, Bycroft M. Structure of the Jab1/MPN domain and its implications for proteasome function. Biochemistry 2003;42:11460–5
- Lingaraju GM, Bunker RD, Cavadini S, et al. Crystal structure of the human COP9 signalosome. Nature 2014;512:161–5
- Infante P, Mori M, Alfonsi R, et al. Gli1/DNA interaction is a druggable target for Hedgehog-dependent tumors. EMBO J 2015;34:200–17
- Mascarello A, Mori M, Chiaradia-Delatorre LD, et al. Discovery of Mycobacterium tuberculosis protein tyrosine phosphatase B (PtpB) inhibitors from natural products. PLoS One 2013;8:e77081
- Congreve M, Carr R, Murray C, Jhoti H. A ‘rule of three’ for fragment-based lead discovery? Drug Discov Today 2003;8:876–7
- Day JA, Cohen SM. Investigating the selectivity of metalloenzyme inhibitors. J Med Chem 2013;56:7997–8007
- FRED, version 3.0.1. OpenEye scientific coftware, Santa Fe, NM. Available from: http://www.eyesopen.com [last accessed 25 Feb 2016]
- McGann M. FRED pose prediction and virtual screening accuracy. J Chem Inf Model 2011;51:578–96
- McGann M. FRED and HYBRID docking performance on standardized datasets. J Comput Aided Mol Des 2012;26:897–906
- Licursi V, Salvi C, De Cesare V, et al. The COP9 signalosome is involved in the regulation of lipid metabolism and of transition metals uptake in Saccharomyces cerevisiae. FEBS J 2014;281:175–90
- Nielsen TK, Hildmann C, Dickmanns A, et al. Crystal structure of a bacterial class 2 histone deacetylase homologue. J Mol Biol 2005;354:107–20
- Holmes IP, Gaines S, Watson SP, et al. The identification of β-hydroxy carboxylic acids as selective MMP-12 inhibitors. Bioorg Med Chem Lett 2009;19:5760–3
- Bertini I, Calderone V, Fragai M, et al. X‐ray structures of binary and ternary enzyme‐product‐inhibitor complexes of matrix metalloproteinases. Angew Chem 2003;115:2777–80
- Mori M, Massaro A, Calderone V, et al. Discovery of a new class of potent MMP inhibitors by structure-based optimization of the arylsulfonamide scaffold. ACS Med Chem Lett 2013;4:565–9
- Panina NS, Belyaev AN, Simanova SA. Carboxylic acids and their anions. Acid and ligand properties. Russ J Gen Chem 2002;72:91–4
- Rinaldi T, Ricci C, Porro D, et al. A mutation in a novel yeast proteasomal gene, RPN11/MPR1, produces a cell cycle arrest, overreplication of nuclear and mitochondrial DNA, and an altered mitochondrial morphology. MBC 1998;9:2917–31
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970;227:680–5
- Shivakumar D, Williams J, Wu Y, et al. Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force field. J Chem Theory Comput 2010;6:1509–19
- Bowers KJ, Chow E, Xu H, et al. Scalable algorithms for molecular dynamics simulations on commodity clusters. Conference, Proceedings of the ACM/IEEE on Supercomputing, 2006, p. 43
- OEDocking 3.0.1: OpenEye Scientific Software, Santa Fe, NM. Available from: http://www.eyesopen.com [last accessed 25 Feb 2016]
- FILTER 2.5.1.4: OpenEye Scientific Software, Santa Fe, NM. Available from: http://www.eyesopen.com [last accessed 25 Feb 2016].
- QUACPAC 1.5.0: OpenEye Scientific Software, Santa Fe, NM. Available from: http://www.eyesopen.com [last accessed 25 Feb 2016]
- OMEGA2 2.4.6: OpenEye Scientific Software, Santa Fe, NM. Available from: http://www.eyesopen.com [last accessed 25 Feb 2016]
- Hawkins PC, Skillman AG, Warren GL, et al. Conformer generation with OMEGA: algorithm and validation using high quality structures from the Protein Databank and Cambridge Structural Database. J Chem Inf Model 2010;50:572–84
- MarvinSketch 6.1.4: ChemAxon. Available from: http://www.chemaxon.com [last accessed 25 Feb 2016]
- Case DA, Darden TA, Cheatham TE, et al. AMBER 12; University of California, San Francisco, 2012
