Abstract
Cancer multidrug resistance (MDR) occurs when cancer cells evade the cytotoxic actions of chemotherapeutics through the active efflux of drugs from within the cells. Our group have previously demonstrated that multidrug-resistant breast cancer cells spontaneously shed microparticles (MPs) and that these MPs can transfer resistance to drug-responsive cells and confer MDR on those cells in as little as 4 h. Furthermore, we also showed that, unlike MPs derived from leukaemia cells, breast cancer–derived MPs display a tissue selectivity in the transfer of P-glycoprotein (P-gp), transferring the resistance protein only to malignant breast cells. This study aims to define the proteome of breast cancer–derived MPs in order to understand the differences in protein profiles between those shed from drug-resistant versus drug-sensitive breast cancer cells. In doing so, we detail the protein cargo required for the intercellular transfer of MDR to drug-sensitive recipient cells and the factors governing the transfer selectivity to malignant breast cells. We describe the first proteomic analysis of MPs derived from human breast cancer cells using SDS PAGE and liquid chromatography–tandem mass spectrometry (LC/MS/MS), in which we identify 120 unique proteins found only in drug-resistant, breast cancer–derived MPs. Our results demonstrate that the MP-mediated transfer of P-gp to recipient cells occurs alongside CD44; the Ezrin, Radixin and Moesin protein family (ERM); and cytoskeleton motor proteins within the MP cargo.
To access the supplementary material to this article, please see Supplementary files under Article Tools online.
The major obstacle in the successful treatment of breast cancer is the ability of tumour cells to resist drugs through the acquisition of multidrug resistance (MDR). MDR is a unique type of drug resistance resulting in cross-resistance to a wide array of structurally and functionally unrelated drugs following exposure to a single chemotherapeutic (Citation1). This capacity is attributed to the overexpression of efflux proteins belonging to the ABC superfamily of membrane transporters (Citation2). Transporters responsible for MDR include P-glycoprotein (P-gp) (Citation3) and the multidrug resistance–associated protein (MRP1) (Citation4).
Cells communicate and exchange molecules with each other either by direct cell-to-cell contact (Citation5) or through the dissemination of microvesicles, including microparticles (MPs) (Citation6), exosomes (Citation7), apoptotic bodies (Citation8), tunnelling nanotubes (Citation9) and/or filopodial bridges (Citation10). During intercellular communication, cells transfer nucleic acids and proteins, which subsequently affect the phenotype of the recipient cells (Citation11). Our laboratory was the first to show that P-gp can be disseminated within a cancer cell population and acquired by drug-sensitive cells through a process of intercellular binding and transfer mediated by MPs (Citation6, Citation11, Citation12). This mechanism allows for the intercellular dissemination of cancer MDR both in vitro and in vivo (Citation13). Furthermore, we recently showed that breast cancer–derived MPs display a tissue selectivity in the transfer of P-gp, transferring the resistance protein to malignant breast cells only (Citation13).
MPs are small membrane vesicles arising from membrane budding and are observed in most cell types during apoptosis or upon cell activation (Citation14). MPs serve an important role in physiology, specifically in maintaining haemostasis (Citation15). They are also mediators in various disease states, including hypercoagulation and thromboembolism (Citation16), inflammation (Citation17), diabetes (Citation18), HIV-1 (Citation19) and cancer (Citation20).
Interaction between proteins associated with the plasma membrane and the cytoskeleton is crucial in cell adhesion, cell signalling, membrane trafficking, cell motility, apoptosis and MP formation (Citation21–Citation23). A family of closely related proteins, the ERM (Ezrin, Radixin and Moesin) proteins, play an extensive role in these processes and possess a unique module required to interface and crosslink membrane proteins with the cytoskeleton (Citation21, Citation23).
This research defines the proteome of breast cancer–derived MPs and compares the protein content of MPs shed from drug-sensitive cells with their drug-resistant counterparts. In doing so, we establish the protein cargo required for the intercellular dissemination of MDR by MPs and identify the protein signature associated with the selectivity of P-gp transfer by resistant breast cancer–derived MPs.
Materials and methods
Cell culture
The drug-sensitive human breast adenocarcinoma cell line MCF-7 and the drug-resistant cell line MCF-7/Dx were used in these studies. The cell lines have been validated as appropriate models for the study of P-gp-mediated MDR in vitro and in vivo (Citation13). Cells were cultured in RPMI 1640 medium (Sigma-Aldrich), supplemented with 10% (v/v) heat-inactivated foetal bovine serum (Life Technologies) and maintained in the absence of antibiotics at 37°C and 5% CO2.
Harvesting MPs
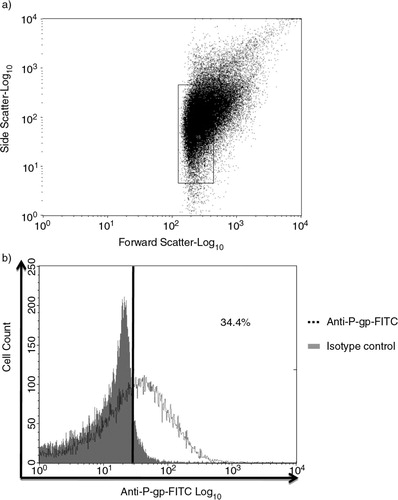
We have extensively described the isolation and validation of the MP population for morphology, size and resistance protein expression previously, using scanning electron microscope (Citation24, Citation25) and flow cytometric analysis (FCM) (Citation6, Citation13, Citation24–Citation26). Likewise in this study, MPs were gated based on size (). Thirty-four percent of the gated population detected positive for P-gp expression using an antibody selective for the external epitope. This is consistent with our previous findings for these MPs (Citation13, Citation25). The procedure for isolation of MPs was as described by us in Refs. (Citation6, Citation13, Citation24–Citation26). In brief, after the cells have reached confluence, the supernatant containing cells and MPs was collected and centrifuged for 5 min at 500 g to remove cell and cell debris. The supernatant was further centrifuged for 5 min at 500 g and 1 h at 15,000 g at 17°C to pellet MPs. The pellet was washed in serum-free media and centrifuged for 2 min at 2,000 g to remove debris. The media containing MPs were further centrifuged for 30 min at 18,000 g at 17°C, and the pellet was resuspended in serum-free RPMI-1640 and/or low-volume of alkaline extraction reagent (pH 8.8) containing 7 M urea, 2 M thiourea, 50 mM tris(hydroxymethyl)methylamine-HCl and 1% C7BzO in a 10 mL final volume of water (Citation27). MPs resuspended in serum-free RPMI-1640 were validated by FCM (LSRII, BD Biosciences, CA, USA) by P-gp (clone 17F9, BD Biosciences, Australia) staining as described in Ref. (Citation25).
Fig. 1 Characterization of microparticles (MPs) isolated from MCF-7/Dx cells. a) MPs isolated from MCF-7/Dx cells were analysed via flow cytometry and gated based on sized latex beads (0.3–1.1 µM); b) 34.4% of the gated MP population detected positive for P-gp expression using anti-P-gp mAb (BD Bioscience). All experiments were repeated at least 3 times with similar results. Data are representative of a typical experiment.
Protein extraction
The pellet, resuspended in a low volume of alkaline extraction, was sonicated 4 times for 30 sec at 40% power, using a high-intensity ultrasonic processor (50 watt, Sonics and Materials Inc., USA). The samples were reduced and alkylated in the alkaline protein solution by adding tributylphosphine solution to a final concentration of 5 mM (1:40 dilution) and acrylamide monomers to a final concentration of 20 mM (1:50 dilution). The reaction was quenched by adding 1 M dithiothreitol solution (final concentration: 20 mM) and centrifuged at 17,000 g for 5 min to pellet any undissolved material. Finally, the protein was precipitated with 5 volumes of acetone before resuspension in 1% SDS (sodium dodecyl sulphate). Total protein concentration was determined using a Qubit fluorometer (Invitrogen) as per the manufacturer's protocol.
SDS-PAGE and trypsin in-gel digestion
One hundred micrograms of extracted MP proteins was mixed with 2× SDS sample buffer containing Tris-HCl, glycerol, SDS, bromophenol blue and water (pH 8.8) at the ratio of 2:1 (protein–SDS buffer), and sonicated in a water-bath for 5 min followed by centrifugation for 5 min at 16,500 g, room temperature. Proteins were separated using a 4–10% gel at a voltage of 150 V in MES SDS running buffer (Invitrogen). The gel was fixed with 40% methanol–10% acetic acid for 30 min with gentle shaking before staining with Coomassie blue overnight. In-gel trypsin digestion was performed as described in Ref. (Citation27). Briefly, the gel (Supplementary file 1) was sectioned according to the MW markers into 12 different sections, and the sections diced into 1×1 mm pieces which were then de-stained by adding 50% acetonitrile (ACN)–50 mM NH4HCO3 and incubating for 10 min at room temperature. The process was repeated until the colour disappeared. One hundred percent ACN was added to dehydrate the gel pieces before rehydration with 12.5 ng/µL of trypsin solution (Trypsin gold-MS grade, Promega) and overnight incubation at 37°C. After incubation, the supernatant was collected after sonication in a water-bath for 10 min, followed by another sonication after the addition of 30 µL of 50% ACN–2% formic acid. The solution was added to the previously collected peptides, and the volume reduced to 15 µL by rotary evaporation. The peptide solution was centrifuged at 14,000 g for 10 min to remove any insoluble material prior to LC/MS/MS analysis.
Western blotting
Twenty micrograms of total cellular and MP proteins were separated on SDS-PAGE, as described earlier, before transferring to PVDF membrane (Pall Australia, VIC, Australia). The membrane was blocked overnight with 5% skim milk in PBS and 0.05% Tween 20, and then incubated with anti-P-gp mAb (clone F4; Sigma-Aldrich), anti-Ezrin mAb (clone 3C12; Life Science), anti-Moesin mAb (clone 38/87; Sigma-Aldrich), anti-radixin pAb (SAB2500859; Sigma-Aldrich) or CD44 mAb (clone EPR1013Y; Abcam) for 1 h. Anti-β-actin (clone AC-74; Sigma-Aldrich) was used as the internal control, followed by horseradish peroxidase conjugates secondary antibody. Protein expression was visualized using an ECL (enhanced chemoluminescence) system (Roche Applied Science, NSW, Australia). The membranes were imaged using the luminescent image analyser LAS-3000 (Fujifilms, Brookvale, NSW, Australia).
LC/MS/MS
Using an Eksigent AS-1 autosampler connected to a Tempo nanoLC system (Eksigent, USA), 10 µL of the sample was loaded at 20 µL/min with MS loading solvent (2% ACN+0.2% trifluoroacetic acid) onto a C8 trap column (CapTrap, Michrom Biosciences, USA). After washing the trap for 3 min, the peptides were washed off the trap at 300 nL/min with MS solvent A (2% ACN+0.2% formic acid) onto a PicoFrit column (75×150 mm, New Objective) packed with Magic C18AQ resin (Michrom Biosciences). Peptides were eluted from the column and into the source of a QSTAR Elite hybrid quadrupole-time-of-flight mass spectrometer (ABSciex) using the following program: 5–50% MS solvent B (98% ACN+0.2% formic acid) over 30 min, 50–80% MS solvent B over 5 min, 80% MS solvent B for 2 min and 80–5% for 3 min. The eluting peptides were ionized from the PicoFrit column at 2300 V. An Intelligent Data Acquisition (IDA) experiment was performed, with a mass range of 350–1,500 Da continuously scanned for peptides of charge state 2+–5+ with an intensity of more than 30 counts/s. Selected peptides were fragmented, and the product ion fragment masses measured over a mass range of 100–1,500 Da. The mass of the precursor peptide was then excluded for 15 sec.
Data analysis
Three biological replicates of total MP proteins were analysed by LC-MS/MS, and the MS/MS data files produced by the QSTAR Elite were searched using Mascot Daemon (version 2.3, provided by the Walter and Eliza Hall Institute, (Citation28)) and searched against the LudwigNR database (composed of the UniProt, plasmoDB and Ensembl databases (vQ213)) with the following parameter settings. Taxonomy: Homo sapiens. Fixed modifications: none. Variable modifications: propionamide, oxidized methionine and deamidated asparagine. Enzyme: semi-trypsin. Number of allowed missed cleavages: 3. Peptide mass tolerance: 100 ppm. MS/MS mass tolerance: 0.2 Da. Charge state: 2+, 3+ and 4+. Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm (Citation29). Protein identifications were accepted if they could be established at greater than 95.0% probability and contained at least 2 identified peptides in 2 replicates. Protein probabilities were assigned by the Protein Prophet algorithm (Citation30). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were clustered by Scaffold to satisfy the principles of parsimony.
Results and discussion
The production and secretion of extracellular vesicles or MPs are universal cellular processes that have been described in bacteria, archaea and eukaryotes (Citation31, Citation32). MPs package proteins, DNA and RNA, and they display a variety of cell surface markers. These features affect how MPs identify target cells and the composition of the cargo they transport to sites distal from their origin (Citation33). In higher eukaryotes, MPs have a range of pathophysiological roles in cancer, infectious disease, systemic lupus, arthritis and neurodegenerative disorders, and their presence has been detected in most body fluids (Citation31, Citation34). Because of their informational content, MPs have been described as signalosomes (Citation31). MPs are involved in tumour spread (Citation35) and trigger tumour growth by stimulating the proliferation of cancerous cells by inducing angiogenesis in neighbouring endothelial cells (Citation36). Previous work in our laboratory has shown for the first time that purified MPs transfer functional P-gp from drug-resistant cancer cells to drug-sensitive cells (Citation6). We were able to show that MPs can confer mRNA, which has implications in the transfer and dominance of cancer traits (Citation11, Citation24). In breast cancer, MPs show tissue selectivity by transferring P-gp only to malignant breast cancer cells but not to other cells (Citation13). We set out to profile the proteome of MPs derived from breast cancer cells to identify proteins involved in MDR transfer. Thus, in this study, we applied a conceptually unbiased proteomic approach to interrogate the protein content of MPs purified from isogenic breast cancer–derived lines that differ in their resistance profile from a broad range of drugs. Initially, more than 600 proteins were identified in both samples. Applying a stringent criterion for protein validation of a minimum of 2 peptides in at least 2 technical replicates reduced the final analysis to 270 total proteins. Of the 270, 249 proteins belong to MCF-7/Dx-MPs and 150 proteins belong to MCF-7-MPs, with 129 common to both (, Supplementary file 2). The unique proteins identified in this experiment were also consistent with the Western blot data shown in . We observed a selective packaging of ERM and CD44 proteins in MCF-7/Dx-MPs relative to MCF-7-MPs consistent with our LC-MS/MS data (). Our findings with ezrin are consistent with our previous study that ezrin is present in MP cargo isolated from resistant cells (Citation13). In the context of breast cancer, the selective packaging of these proteins in MCF-7/Dx-MPs and not in MCF-7-MPs suggests that these cytoskeletal elements are required in stabilizing P-gp in the MP cargo en route to the recipient cells. This is consistent with previous publications, which show a role for the FERM domain-binding proteins and CD44 in P-gp membrane localization (Citation22, Citation52). This finding expands our knowledge of the required protein signature for P-gp transfer by MPs.
Fig. 2 Venn diagram depicting the comparison of proteins identified in microparticles (MPs). Proteins identified in MPs derived from drug-sensitive breast cancer cells (MCF-7) and drug-resistant breast cancer cells (MCF-7/Dx).
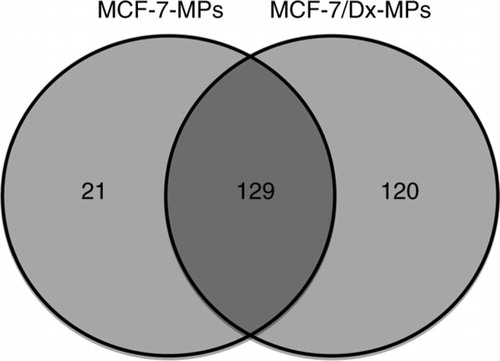
Fig. 3 Western blot analysis of selectively packaged proteins in MCF-7-Dx-MPs (microparticles). Twenty micrograms of total cell and MP lysates of drug-sensitive breast cancer cells (MCF-7) and drug-resistant breast cancer cells (MCF-7/Dx) were analysed by Western blot to detect (a) P-glycoprotein, 170 kDa; (b) ezrin, ~80 kDa; (c) moesin, 78–80 kDa; (d) CD44, 82 kDa; and (e) radixin, ~80 kDa. Lanes 1–4 are MCF-7 cells, MCF-7-MPs, MCF-7/Dx cells, and MCF-7/Dx-MPs, respectively. β-actin (42 kDa) was used as the internal control. Data are representative of a typical experiment.
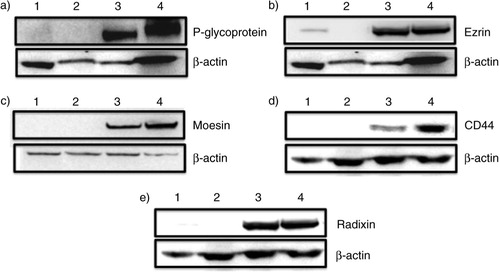
Table I Unique proteins identified in drug-resistant microparticles, which are correlated with cancer and/or multidrug resistance
Gene ontology of the unique proteins identified from drug-resistant MPs was generated according to their annotated (a) cellular component, (b) biological process and (c) molecular function. Besides protein profiling, our interest was to identify candidate proteins that may contribute to the ability of MPs to transfer MDR. The proteins unique to drug-resistant cell MPs were investigated for documented evidence of an association with P-gp and enhancement of drug-resistant activity. Nineteen cytoskeletal proteins, including vimentin, talin-1, coronin 1C and members of the ERM family, were identified. KEGG pathway analysis was able to classify 10 of the unique proteins to the “regulation of actin cytoskeleton” pathway. Those 10 proteins are myosin-9, vinculin, moesin, actin 4, ezrin, integrin beta 3, fibronectin, ras-related C3 botulinum toxin substrate 1 (rho family), IQ motif containing GTPase activating protein 1 and myosin light chain 12B regulatory protein. Most of the cytoskeletal-associated proteins identified possess an N-terminally located FERM domain (4.1 protein, Ezrin, Radixin and Moesin), which plays a key role in linking actin fibres to the plasma membrane. Forty-five proteins related to or known to localize to the plasma membrane were also identified, including multidrug-resistant protein 1, stomatin, aminopeptidase, sarcoplasmic–endoplasmic reticulum calcium ATPase 2, ecto-5′-nucleotidase and glutathione S-transferase P1 (GSTP1). Ecto-5′-nucleotidase (CD73), a membrane-associated glycoprotein that generates adenosine by hydrolysing 5′-AMP to adenosine, has been implicated in metastasis-promoting activities, including tumour angiogenesis and vasodilation, adhesion of lymphocytes to endothelial cells and adhesion, invasion and migration of human breast cancer cells (Citation38, Citation53, Citation54). Increased levels of CD73 protein expression were reported in several MDR cell lines, suggesting that this enzyme is involved in drug resistance (Citation39, Citation55). Expression of various isoforms of glutathione S–transferase P1 is enhanced in adenocarcinoma cells during their transition to a drug-resistant state (Citation40). Involvement of these proteins is likely to play an important role in intracellular trafficking of P-gp within recipient cells because P-gp is localized not only in the plasma membrane but also in intracellular compartments, including the endoplasmic reticulum, golgi, endosomes and lysosomes (Citation56).
The FERM domain is found in a number of cytoskeletal-associated proteins, including Band 4.1, ezrin, radixin, moesin, DAL-1, talin-1, merlin, focal adhesion kinase (FAK) and the proline-rich tyrosine kinase 2 (PYK2) (Citation41). This specialized group of macromolecules is found in higher order structures, including sites of cell–cell and cell–extracellular matrix attachment (Citation42, Citation57). Ezrin, radixin, moesin and talin-1 were found in MPs derived from drug-resistant cells only. The N-terminally located FERM domain binds to cytosolically exposed C-terminal regions of integral membrane proteins, and C-terminal domains in FERM proteins bind actin (Citation43, Citation58). Interactions between the plasma membrane and the cytoskeleton are essential in cell adhesion, cell signalling, membrane trafficking, cell motility, apoptosis and MP formation (Citation21–Citation23). The presence of FERM domain proteins in drug-resistant MPs indicates that they are likely to play a key role in the mechanism by which MPs transfer an MDR phenotype.
There are several key observations that support the contention that MDR involves changes to cytoskeletal structure. These include (a) the observation that MDR osteosarcoma cells exhibit an increase in actin–stress fibre organization which, when disrupted, sensitizes cells to anticancer drugs (Citation59); (b) P-gp translocates to the plasma membrane and co-localizes with ezrin in IFN-γ treated, monocyte-derived macrophages (Citation60); (c) the co-localization of P-gp with ERM proteins occurs on pseudopods and uropods in resistant lymphoid cells (Citation22), and disruption of the ERM–P-gp association was shown to impair P-gp function and resulted in the cellular redistribution of P-gp (Citation22); (d) an actin–P-gp interaction is essential for endosomal trafficking of P-gp (Citation61); and (e) P-gp binds ezrin at amino acid residues 149–242 in the FERM domain, and the disruption of this interaction results in P-gp relocation from the plasma membrane to the cytoplasm (Citation62).
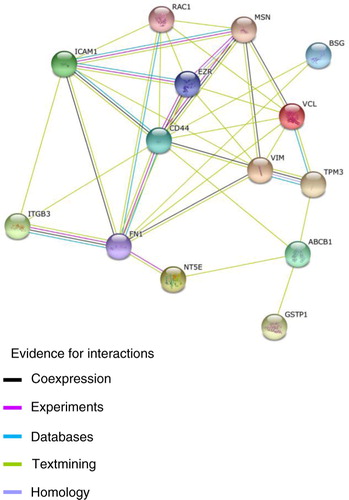
CD44, stathmin 1 and fermitin family homolog 2 (FERMT2) proteins were identified exclusively in MCF-7/Dx-MPs. CD44 is the major surface receptor to hyaluronan and is implicated in cell adhesion, metastasis and motility (Citation63, Citation64). C-terminal regions of CD44 located intracellularly assemble with FERM domain proteins, including ezrin, radixin and moesin, that anchor CD44 to actin and influence downstream signalling pathways (Citation37). The ezrin binding site on CD44 has been mapped to 2 clusters of basic amino acids in a membrane-proximal 9-amino-acid region within the CD44 cytoplasmic domain (Citation65). CD44 and P-gp typically co-immunoprecipitate (Citation52), their expression in the cell is co-regulated (Citation66) and both proteins are found co-localized to the plasma membrane (Citation52). Supporting this model of interaction, it has recently been proposed that P-gp complexes with activated CD44 via ERM intermediates (Citation67). Low-molecular-mass hyaluronic acid promotes the internalization of the CD44–ERM protein complex, which is accompanied by decreased drug resistance (Citation37). These observations are supported by the STRING database (), which shows P-gp interacting with CD44 along with GSTP1, tropomyosin 3 and CD73. CD44 also interacts with intercellular adhesion molecule 1 (ICAM 1), rho family, fibronectin 1, ezrin, moesin, integrin beta 3, vinculin, basigin and vimentin. These proteins have a role in promoting cancer cell proliferation and drug resistance. Vinculin modulates talin to help in cell adhesion by mediating integrin-mediated adhesion (Citation46). Integrins are cell surface receptors that participate in cell–cell and cell–extracellular matrix interactions and contribute to the aggressive behaviour of cancer cells (Citation68). We found integrin-β-3 in resistant cell MPs, whereas integrin-β-2 was present in both MP preparations. Integrins are also known to transmit chemical signals and have a role in cellular responses such as migration, survival, differentiation and motility (Citation69). Besides vinculin and talin, the kindlin family proteins also enhance integrin activation (Citation70). FERMT2 proteins, which belong to the kindlin family, were identified exclusively in MCF-7/Dx-MPs.
Fig. 4 Protein network generated from String 9.05. Network of unique proteins associated with ABCB1/P-gp, isolated from microparticles derived from drug-resistant breast cancer cell (MCF-7/Dx), visualized on the String website. Proteins such as CD44, tropomyosin 3, glutathione S-transferase P1 and ecto-5′-nucleotidase are associated with drug-resistant proteins, whereas fibronectin 1, intercellular adhesion molecule 1, ezrin, moesin, integrin beta 3, vinculin, basigin, rho family and vimentin are associated with CD44.
Other MCF-7/Dx-specific MP proteins identified in our proteome analysis that have a role in promoting cancer cell proliferation and drug resistance included stathmin, aminopeptidase, phosphoserine aminotransferase and other proteins listed in . Stathmin has a role in cell proliferation and participates in carcinogenesis of various carcinomas (Citation48), and aminopeptidases are reported to play a role in tumour invasion and are found to be elevated in the plasma and effusion of cancer patients (Citation71), whereas phosphoserine aminotransferase is reported to stimulate cell growth and increases the chemotherapy resistance of colon cancer cells (Citation47).
Conclusion
Previously, we have reported that resistance to anticancer drugs can be transferred by MPs from drug-resistant to drug-sensitive cells within a cancer cell population (Citation6, Citation11, Citation12) in a tissue-selective manner (Citation13). In this work, we characterize the proteome of MPs derived from drug-resistant versus drug-sensitive breast cancer cells and show that the proteome of MCF-7/Dx-MPs has an abundance of proteins previously reported to be involved in MDR. These findings reinforce our earlier observations and suggest that the transfer of MDR and specifically P-gp to recipient cells is facilitated by cytoskeletal proteins, actin binding proteins, glycolytic proteins, adhesion molecules and cytoskeletal motor proteins (, ). These findings bring us closer to identifying targets, which may be used in the design of selective therapeutics for the prevention of MDR clinically.
Fig. 5 Microparticles (MPs) play a deleterious role in cancer by transferring drug-resistant proteins. MP surface molecules (adhesion molecules) and FERM domain proteins are proposed to be associated with tissue selectivity in the transfer of P-gp in malignant breast cells.
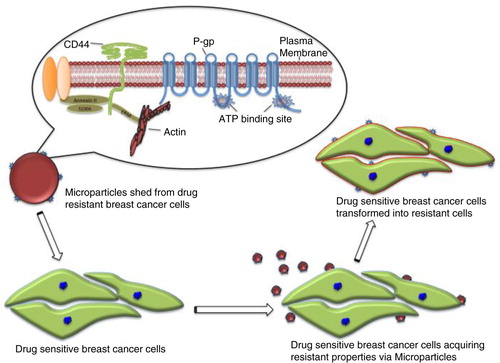
Table II Pathway analysis of proteins unique to resistant breast cancer–derived microparticles
Authors’ contributions
Deep Pokharel: performed experiments, analysed data and prepared the manuscript; Matthew P. Padula: technical support and advice, conceptual advice and revised the manuscript; Jamie F. Lu: flow cytometry analysis and revised the manuscript; Jessica L. Tacchi: technical support and advice, conceptual advice and revised the manuscript; Frederick Luk: technical support and advice, conceptual advice and revised the manuscript; Steven P. Djordjevic: technical advice, conceptual advice and revised the manuscript; and Mary Bebawy: designed the research project, conceptual advice and revised the manuscript.
Conflict of interest and funding
The authors have not received any funding or benefits from industry or elsewhere to conduct this study.
Supplementary Material
Download PDF (406.1 KB)Acknowledgements
The authors thank the National Health and Medical Research Council (1007613) for research grants to Associate Professor Mary Bebawy.
Notes
To access the supplementary material to this article, please see Supplementary files under Article Tools online.
References
- Biedler JL, Riehm H, Peterson RH, Spengler BA. Membrane-mediated drug resistance and phenotypic reversion to normal growth behavior of Chinese hamster cells. J Natl Cancer Inst. 1975; 55: 671–80.
- Fojo A, Akiyama S, Gottesman MM, Pastan I. Reduced drug accumulation in multiply drug-resistant human KB carcinoma cell lines. Cancer Res. 1985; 45: 3002–7.
- Riordan JR, Deuchars K, Kartner N, Alon N, Trent J, Ling V. Amplification of P-glycoprotein genes in multidrug-resistant mammalian cell lines. Nature. 1985; 316: 817–9. 10.1038/316817a0.
- Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP–dependent transporters. Nat Rev Cancer. 2002; 2: 48–58.
- Levchenko A, Mehta BM, Niu X, Kang G, Villafania L, Way D et al. Intercellular transfer of P-glycoprotein mediates acquired multidrug resistance in tumor cells. Proc Natl Acad Sci USA. 2005; 102: 1933–8.
- Bebawy M, Combes V, Lee E, Jaiswal R, Gong J, Bonhoure A et al. Membrane microparticles mediate transfer of P-glycoprotein to drug sensitive cancer cells. Leukemia. 2009; 23: 1643–9.
- Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007; 9: 654–9.
- Bergsmedh A, Szeles A, Henriksson M, Bratt A, Folkman MJ, Spetz A-L et al. Horizontal transfer of oncogenes by uptake of apoptotic bodies. Proc Natl Acad Sci USA. 2001; 98: 6407–11.
- Watkins SC, Salter RD. Functional connectivity between immune cells mediated by tunneling nanotubules. Immunity. 2005; 23: 309–18.
- Sherer NM, Mothes W. Cytonemes and tunneling nanotubules in cell–cell communication and viral pathogenesis. Trends Cell Biol. 2008; 18: 414–20.
- Jaiswal R, Gong J, Sambasivam S, Combes V, Mathys J-M, Davey R et al. Microparticle-associated nucleic acids mediate trait dominance in cancer. FASEB J. 2012; 26: 420–9.
- Bebawy M, Morris MB, Roufogalis BD. A continuous fluorescence assay for the study of P-glycoprotein-mediated drug efflux using inside-out membrane vesicles. Anal Biochem. 1999; 268: 270–7.
- Jaiswal R, Luk F, Dalla PV, Grau GER, Bebawy M. Breast cancer-derived microparticles display tissue selectivity in the transfer of resistance proteins to cells. PLoS One. 2013; 8: e61515.
- Distler JH, Pisetsky DS, Huber LC, Kalden JR, Gay S, Distler O. Microparticles as regulators of inflammation: novel players of cellular crosstalk in the rheumatic diseases. Arthritis Rheum. 2005; 52: 3337–48.
- Gong J, Jaiswal R, Mathys J-M, Combes V, Grau G, Bebawy M. Microparticles and their emerging role in cancer multidrug resistance. Cancer Treat Rev. 2012; 38: 226–34.
- Zwicker JI, Liebman HA, Neuberg D, Lacroix R, Bauer KA, Furie BC et al. Tumor-derived tissue factor–bearing microparticles are associated with venous thromboembolic events in malignancy. Clin Cancer Res. 2009; 15: 6830–40.
- Vasina E, Heemskerk JW, Weber C, Koenen RR. Platelets and platelet-derived microparticles in vascular inflammatory disease. Inflamm Allergy Drug Targets. 2010; 9: 346–54.
- Sabatier F, Darmon P, Hugel B, Combes V, Sanmarco M, Velut J-G et al. Type 1 and type 2 diabetic patients display different patterns of cellular microparticles. Diabetes. 2002; 51: 2840–5.
- Mack M, Kleinschmidt A, Brühl H, Klier C, Nelson PJ, Cihak J et al. Transfer of the chemokine receptor CCR5 between cells by membrane-derived microparticles: a mechanism for cellular human immunodeficiency virus 1 infection. Nat Med. 2000; 6: 769–75.
- Nieuwland R, van der Post JA, Lok CA, Kenter G, Sturk A. Microparticles and exosomes in gynecologic neoplasias. Semin Thromb Hemost. 2010; 36: 925–9.
- Jeon S, Kim NH, Kim JY, Lee AY. Stem cell factor induces ERM proteins phosphorylation through PI3K activation to mediate melanocyte proliferation and migration. Pigment Cell Melanoma Res. 2009; 22: 77–85.
- Luciani F, Molinari A, Lozupone F, Calcabrini A, Lugini L, Stringaro A et al. P-glycoprotein–actin association through ERM family proteins: a role in P-glycoprotein function in human cells of lymphoid origin. Blood. 2002; 99: 641–8.
- Killock DJ, Parsons M, Zarrouk M, Ameer-Beg SM, Ridley AJ, Haskard DO et al. In vitro and in vivo characterization of molecular interactions between calmodulin, ezrin/radixin/moesin, and L-selectin. J Biol Chem. 2009; 284: 8833–45.
- Lu JF, Luk F, Gong J, Jaiswal R, Grau GE, Bebawy M. Microparticles mediate MRP1 intercellular transfer and the re-templating of intrinsic resistance pathways. Pharmacol Res. 2013; 76: 77–83.
- Gong J, Luk F, Jaiswal R, George AM, Grau GER, Bebawy M. Microparticle drug sequestration provides a parallel pathway in the acquisition of cancer drug resistance. Eur J Pharmacol. 2013; 721: 116–25.
- Jaiswal R, Luk F, Gong J, Mathys J-M, Grau G, Bebawy M. Microparticle conferred microRNA profiles-implications in the transfer and dominance of cancer traits. Mol Cancer. 2012; 11: 37.
- Bogema DR, Deutscher AT, Woolley LK, Seymour LM, Raymond BB, Tacchi JL et al. Characterization of cleavage events in the multifunctional cilium adhesin Mhp684 (P146) reveals a mechanism by which Mycoplasma hyopneumoniae regulates surface topography. MBio. 2012; 3: e00282–11.
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999; 20(18): 3551–67.
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002; 74: 5383–92.
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometrym. Anal Chem. 2003; 75: 4646–58.
- Andaloussi SE, Mäger I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013; 12: 347–57.
- Lloubes R, Bernadac A, Pommier S. Non classical secretion systems. Res Microbiol. 2013; 164: 655–63.
- Lee Y, Andaloussi SE, Wood MJ. Exosomes and microvesicles: extracellular vesicles for genetic information transfer and gene therapy. Hum Mol Genet. 2012; 21: R125–34.
- Kucharzewska P, Belting M. Emerging roles of extracellular vesicles in the adaptive response of tumour cells to microenvironmental stress. J Extracell Vesicles. 2013; 2 20304, doi: http://dx.doi.org/10.3402/jev.v2i0.20304.
- Dvorak H, Quay S, Orenstein N, Dvorak A, Hahn P, Bitzer A et al. Tumor shedding and coagulation. Science. 1981; 212: 923–4.
- Al-Nedawi K, Meehan B, Kerbel RS, Allison AC, Rak J. Endothelial expression of autocrine VEGF upon the uptake of tumor-derived microvesicles containing oncogenic EGFR. Proc Natl Acad Sci USA. 2009; 106: 3794–9.
- Zöller M. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule?. Nat Rev Cancer. 2011; 11: 254–67.
- Wang L, Zhou X, Zhou T, Ma D, Chen S, Zhi X et al. Ecto-5'-nucleotidase promotes invasion, migration and adhesion of human breast cancer cells. J Cancer Res Clin Oncol. 2008; 134: 365–72.
- Ujhazy P, Berleth ES, Pietkiewicz JM, Kitano H, Skaar JR, Ehrke MJ et al. Evidence for the involvement of ecto-5'-nucleotidase (CD73) in drug resistance. Int J Cance. 1996; 68: 493–500.
- Kalinina E, Berozov T, Shtil A, Chernov N, Glasunova V, Novichkova M et al. Expression of genes of glutathione transferase isoforms GSTP1-1, GSTA4-4, and GSTK1-1 in tumor cells during the formation of drug resistance to cisplatin. Bull Exp Biol Med. 2012; 154: 64–7.
- Meurice N, Wang L, Lipinski CA, Yang Z, Hulme C, Loftus JC. Structural conservation in band 4.1, ezrin, radixin, moesin (FERM) domains as a guide to identify inhibitors of the proline-rich tyrosine kinase 2. J Med Chem. 2009; 53: 669–77.
- Amieva MR, Furthmayr H. Subcellular localization of moesin in dynamic filopodia, retraction fibers, and other structures involved in substrate exploration, attachment, and cell-cell contacts. Exp Cell Res. 1995; 219: 180–96.
- Pearson MA, Reczek D, Bretscher A, Karplus PA. Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain. Cell. 2000; 101: 259–70.
- Dixon J, Kaklamanis L, Turley H, Hickson I, Leek R, Harris A et al. Expression of aminopeptidase-n (CD 13) in normal tissues and malignant neoplasms of epithelial and lymphoid origin. J Clin Pathol. 1994; 47: 43–7.
- Kuhajda FP. Fatty-acid synthase and human cancer: new perspectives on its role in tumor biology. Nutrition. 2000; 16: 202–8.
- Critchley D. Cytoskeletal proteins talin and vinculin in integrin-mediated adhesion. Biochem Soc Trans. 2004; 32: 831–6.
- Vié N, Copois V, Bascoul-Mollevi C, Denis V, Bec N, Robert B et al. Overexpression of phosphoserine aminotransferase PSAT1 stimulates cell growth and increases chemoresistance of colon cancer cells. Mol Cancer. 2008; 7: 14.
- Ghosh P, Anderson J, Cohen N, Takeshita K, Atweh G, Lebowitz P. Expression of the leukemia-associated gene, p18, in normal and malignant tissues; inactivation of expression in a patient with cleaved B cell lymphoma/leukemia. Oncogene. 1993; 8: 2869.
- Vadlamudi RK, Li F, Adam L, Nguyen D, Ohta Y, Stossel TP et al. Filamin is essential in actin cytoskeletal assembly mediated by p21-activated kinase 1. Nat Cell Biol. 2002; 4: 681–90.
- Honda K, Yamada T, Endo R, Ino Y, Gotoh M, Tsuda H et al. Actinin-4, a novel actin-bundling protein associated with cell motility and cancer invasion. J Cell Biol. 1998; 140: 1383–93.
- Geiger B, Spatz JP, Bershadsky AD. Environmental sensing through focal adhesions. Nat Rev Mol Cell Biol. 2009; 10: 21–33.
- Miletti-González KE, Chen S, Muthukumaran N, Saglimbeni GN, Wu X, Yang J et al. The CD44 receptor interacts with P-glycoprotein to promote cell migration and invasion in cancer. Cancer Res. 2005; 65: 6660–7.
- Zhi X, Chen S, Zhou P, Shao Z, Wang L, Ou Z et al. RNA interference of ecto-5'-nucleotidase (CD73) inhibits human breast cancer cell growth and invasion. Clin Exp Metastasis. 2007; 24: 439–48.
- Wang L, Tang S, Wang Y, Xu S, Yu J, Zhi X et al. Ecto-5'-nucleotidase (CD73) promotes tumor angiogenesis. Clin Exp Metastasis. 2013; 30: 671–80.
- Rappa G, Lorico A, Flavell RA, Sartorelli AC. Evidence that the multidrug resistance protein (MRP) functions as a co-transporter of glutathione and natural product toxins. Cancer Res. 1997; 57: 5232–7.
- Fu D, Bebawy M, Kable EP, Roufogalis BD. Dynamic and intracellular trafficking of P-glycoprotein-EGFP fusion protein: implications in multidrug resistance in cancer. Int J Cancer. 2004; 109: 174–81.
- Carreiras F, Lehmann M, Sichel F, Marvaldi J, Gauduchon P, Talaer L. Implication of the αvβ3 integrin in the adhesion of the ovarian-adenocarcinoma cell line IGROV1. Int J Cancer. 1995; 63: 530–6.
- Chishti AH, Kim AC, Marfatia SM, Lutchman M, Hanspal M, Jindal H et al. The FERM domain: a unique module involved in the linkage of cytoplasmic proteins to the membrane. Trends Biochem Sci. 1998; 23: 281–2.
- Takeshita H, Kusuzaki K, Ashihara T, Gebhardt MC, Mankin HJ, Hirasawa Y. Actin organization associated with the expression of multidrug resistant phenotype in osteosarcoma cells and the effect of actin depolymerization on drug resistance. Cancer Lett. 1998; 126: 75–81.
- Puddu P, Fais S, Luciani F, Gherardi G, Dupuis ML, Romagnoli G et al. Interferon-gamma up-regulates expression and activity of P-glycoprotein in human peripheral blood monocyte-derived macrophages. Lab Invest. 1999; 79: 1299–309.
- Fu D, Roufogalis BD. Actin disruption inhibits endosomal traffic of P-glycoprotein-EGFP and resistance to daunorubicin accumulation. Am J Physiol Cell Physiol. 2007; 292: C1543–52.
- Brambilla D, Zamboni S, Federici C, Lugini L, Lozupone F, Milito AD et al. P-glycoprotein binds to ezrin at amino acid residues 149–242 in the FERM domain and plays a key role in the multidrug resistance of human osteosarcoma. Int J Cancer. 2012; 130: 2824–34.
- Birch M, Mitchell S, Hart IR. Isolation and characterization of human melanoma cell variants expressing high and low levels of CD44. Cancer Res. 1991; 51: 6660–7.
- Bartolazzi A, Peach R, Aruffo A, Stamenkovic I. Interaction between CD44 and hyaluronate is directly implicated in the regulation of tumor development. J Exp Med. 1994; 180: 53–66.
- Legg JW, Isacke CM. Identification and functional analysis of the ezrin-binding site in the hyaluronan receptor, CD44. Curr Biol. 1998; 8: 705–8.
- Colone M, Calcabrini A, Toccacieli L, Bozzuto G, Stringaro A, Gentile M et al. The multidrug transporter P-glycoprotein: a mediator of melanoma invasion?. J Invest Dermatol. 2007; 128: 957–71.
- Jaiswal R, Raymond Grau GE, Bebawy M. Cellular communication via microparticles: role in transfer of multidrug resistance in cancer. Future Oncol. 2014; 10: 655–69.
- Mizejewski GJ. Role of integrins in cancer: survey of expression patterns. Proc Soc Exp Biol Med. 1999; 222: 124–38.
- Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002; 110: 673–87.
- Montanez E, Ussar S, Schifferer M, Bösl M, Zent R, Moser M et al. Kindlin-2 controls bidirectional signaling of integrins. Genes Dev. 2008; 22: 1325–30.
- Saiki I, Yoneda J, Azuma I, Fujii H, Abe F, Nakajima M et al. Role of aminopeptidase N (CD13) in tumor-cell invasion and extracellular matrix degradationm. Int J Cancer. 1993; 54: 137–43.
