Abstract
Our previous studies have showed that Gli2 played a predominant role in proliferation and apoptosis resistance to TRAIL in hepatoma cells. The purpose of this study was to explore whether Gli2 silencing enhances efficiency of TRAIL for hepatoma in vivo. SMMC-7721-shRNA cells were implanted subcutaneously into nude mices and TRAIL was injected into the peritoneal space. TUNEL assay was used to detect apoptosis of tumor cells. The expression of Gli2, c-FLIPL, c-FLIPS, and Bcl-2 protein was determined by immunohistochemistry, respectively. Apoptosis and the level of caspases proteins in SMMC-7721 and HepG2 cells were detected by Flow cytometry and Western blot. Transcriptional activity of c-FLIP induced by Gli2 was measured by luciferase reporter gene assay. The results showed that lower volumes and weights of tumor were found in mice xenografted with SMMC-7721-shRNA cells as compared with control cells in the presence of TRAIL (P < 0.05). TUNEL assay showed significantly higher apoptosis index (AI) in the SMMC-7721-shRNA group than in the control groups (P < 0.05). There were remarkable positive correlations between Gli2 and c-FLIPL, c-FLIPS, Bcl-2 protein expression. Over-expression of c-FLIP or Bcl-2 in HepG2 cells attenuated TRAIL-induced apoptosis via suppression of caspase-8 or caspase-9 activity, respectively. Luciferase reporter gene assay found a regulatory sequence by which Gli2 activated transcription between -1007 to -244 in the c-FLIP promoter region. This study demonstrates that Gli2 showed regulatory activity on transcription of c-FLIP gene, and Gli2 silencing enhances TRAIL-induced apoptosis via down-regulation of c-FLIP and Bcl-2 in human hepatoma cells in vivo.
Abbreviations
| TRAIL | = | tumor necrosis factor (TNF)-related apoptosis-inducing ligand |
| Gli2 | = | glioma-associated oncogene 2 |
| Hh | = | Hedgehog |
| c-FLIP | = | cellular FLICE-inhibitory protein |
| shRNA | = | short hairpin RNA |
| FADD | = | fas-associated protein with death domain |
| PARP | = | poly ADP-ribose polymerase |
| TUNEL | = | Terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling |
Introduction
Hepatoma is a common malignant tumor with low surgical resection but high recurrence and mortality rates. It is sensitive to neither chemotherapy nor radiotherapy. hepatoma is the third leading cause of death worldwide, with one million deaths annually.Citation1,2 Hepatoma is, therefore, a major threat to human being.
The tumor necrosis factor (TNF)-Related apoptosis inducing ligand (TRAIL) is a new member of the TNF superfamily discovered only in the past few years. A good number of in-vitro laboratory studies have demonstrated that TRAIL induced apoptosis in several tpyes of tumor cells without toxicity to normal cells, including hepatoma cells.Citation3-6 This unique biological characteristic bring potential agent for treatment of tumors. However, the most currently available studies have showed that almost all in-vitro hepatoma cell lines were resistant to TRAIL to different extents. Therefore, understanding the underlying mechanisms reponsible for resistance to TRAIL and restoring sensitivity to TRAIL in hepatoma cells have to be achieved before TRAIL can be used in treatment of hepatoma.
The death receptor pathway by which TRAIL induces cell apoptosis can be regulated at multiple protein levels. Thus, death-receptor-based anti-cancer therapy must be administered concomitantly with counter-resistance agents. Good anti-cancer efficacy is possible only if the antagonists against resistance mechanisms, such as anti-apoptotic proteins including XIAP, c-FLIP and Bcl-2, were developed. The sensitivity of cancer cells to TRAIL can be increased by combining TRAIL with compounds which can reduce the expression level of c-FLIP or Bcl-2.Citation7-9 At present, the XIAP antagonist, Smac analog, and the Bcl-2 antagonist, BH3 analog, either alone or in combination have been studied.Citation10-12 However, there is no report on c-FLIP antagonist.
During embryogenesis, the Hedgehog (Hh) signaling pathway is involved in regulation of normal development of liver, but the pathway is not expressed or mildly expressed in mature liver tissue. Multiple studies in recent years have found strong association between abnormal activation of the Hh signaling pathway and occurrence of hepatic cancer, as well as its malignant biological characteristics.Citation13-15 Gli2 is one of the terminal transcription factors in this signal pathway that carries extracellular Hh signal into the cells. By binding to the promoter region of a downstream gene, Gli2 initiates the transcritpion of target genes, including Gli1. Therefore, it has an extremely important role in the Hh signaling pathway.Citation16 Siegelin et al. have reported that KAAD-cyclopamine facilitated TRAIL-mediated apoptosis by upregulating DR5 and downregulating c-FLIP and Bcl-2 in malignant glioma cells.Citation17 Our previous studies have demonstrated that knockdown of endogenous Gli2 expression sensitized the hepatocellular carcinoma cell line SMMC-7721 to TRAIL-induced apoptosis in vitro. The synergistic action was associated with decreased levels of Bcl-2, c-FLIP, and activation of caspase-dependent pathways.Citation18
In the present study, we found that Gli2 silencing enhanced TRAIL-induced apoptosis through down-regulating c-FLIP and Bcl-2 in vivo, thus revealing the central role of Gli2 in the activation of the antiapoptotic factor c-FLIP, which might represent an important mechanism in the maintenance of resistance to apoptosis in hepatoma treated with TRAIL.
Results
Identification of SMMC-7721 cell line in which Gli2 was stably down-regulated
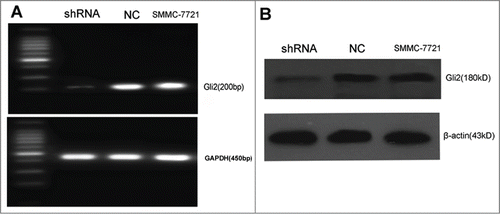
Previous works suggested that downregulation of Gli2 enhanced TRAIL-induced apoptosis in hepatoma cells in vitro. To further investigate whether down-regulation of Gli2 enhances apoptosis of hepatoma cells induced by TRAIL in vivo, we first verified the validity of shRNA that stably downregulated Gli2. As shown in , the mRNA and protein level of Gli2 in SMMC-7721-shRNA cells were markedly decreased as compared with SMMC-7721-NC and SMMC-7721 cells, suggesting that Gli2 was stably down-regulated in SMMC-7721-shRNA cells.
Figure 1. Gli2-specific shRNAs suppressed the expressions of Gli2 mRNA and protein in SMMC-7721 cells. (A) The level of Gli2 mRNA was assessed by semi-quantitative RT-PCR. Total RNA was extracted from SMMC-7721-shRNA, SMMC-7721-NC, SMMC-7721. GAPDH was used as a normalization control. (B) The level of Gli2 protein expression was assessed by Western blot. β-actin was used as a loading control. Densitometric analysis was performed using Quantity one 4.6.2 software. *P < 0.05 compared with control. These experiments were performed in triplicate.
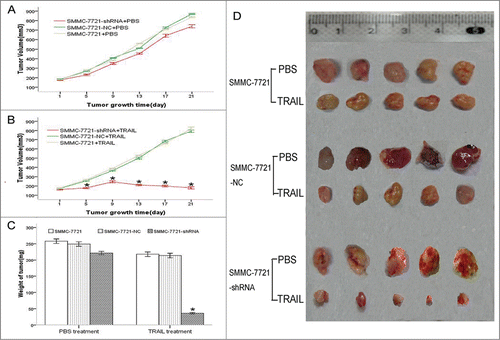
Gli2 gene silencing enhanced growth inhibition of xenograft induced by TRAIL in vivo
In the presence of TRAIL, the primary tumor volumes tremendously decreased in the mice which were xenografted with SMMC-7721-shRNA cells as compared with SMMC-7721-NC cells or untransfected SMMC-7721 cells xenografts. However, there were no significant differences among 3 groups with PBS treatment ().The tumor weights of the 3 groups treated with TRAIL were 36.2 ± 2.12, 244.3 ± 6.57 and 248.5 ± 6.36, respectively (). There were significant differences between the SMMC-7721-shRNA cells and the SMMC-7721-NC cells or the SMMC-7721 cells (P < 0.05), but the differences between the SMMC-7721-NC cells and the SMMC-7721 cells were not significant (P > 0.05). The tumor weights of the 3 groups treated with PBS were 221.2 ± 5.15, 249.1 ± 5.73 and 258.1 ± 6.02, respectively, there were no significant differences. These results indicate that Gli2 gene silencing enhanced growth inhibition of xenograft induced by TRAIL in vivo.
Figure 2. Gli2 gene silencing enhanced growth inhibition of xenograft induced by TRAIL in vivo. (A and B) Growth curves of xenograft in nude mice with PBS or TRAIL treatment. The SMMC-7721-shRNA and the control cells were injected subcutaneously into the BALB/c nude mice at the right flanks. After 1 week, PBS or TRAIL (50 ng/mouse) was injected into the peritoneal space at days 1, 4, 8, 11, 15, and 18, respectively. Tumor size was monitored at intervals of 4 d. (C and D) Weight and Size of tumors in the different groups. On the 21st day after PBS or TRAIL injection, the mice were sacrificed by CO2 inhalation and tumors were resected, weighed (C), and measured (D). Each group consisted of 5 mice. *P < 0.05 compared with control.
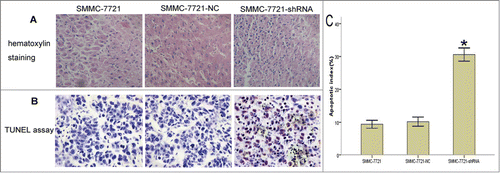
Gli2 gene silencing enhanced TRAIL-induced apoptosis in SMMC-7721 cells in vivo
Since Gli2 gene silencing enhanced growth inhibition of xenograft induced by TRAIL which was proved to induced apoptosis in HCC cells, HE staining and TUNEL assay were used to investigate whether Gli2 effect apoptosis induced by TRAIL in vivo. As compared with SMMC-7721-NC group and the SMMC-7721 group, section from SMMC-7721-shRNA group induced by TRAIL displayed more apoptotic cells with karyopyknosis and red staining of the cytoplasm (). With much more nuclei of cells were stained brown, TUNEL assay showed a high degree of apoptosis in xenografts from SMMC-7721-shRNA group induced by TRAIL, whereas little apoptosis cells were found in xenografts from SMMC-7721-NC or the SMMC-7721 group (). In the presence of TRAIL, theapoptosis index (AI) in xenograft sections was significantly higher in the SMMC-7721-shRNA group than in SMMC-7721-NC or SMMC-7721 group (P < 0.05). However, there was no significant difference of AI between the SMMC-7721-NC and the SMMC-7721 group (, P > 0.05).
Figure 3. Effects of Gli2 silencing on sensitivity of SMMC-7721 cells to TRAIL-induced apoptosis in vivo. After the mice were sacrificed, apoptosis was detected in tumor tissues using HE staining and TUNEL assay. (A) HE staining, magnification × 400. Apoptotic cell showed karyopyknosis and red staining of the cytoplasm. (B) TUNEL assay, magnification × 400. The nucleis of apoptotic cells were stained brown. (C) Bar chart of the apoptosis index (AI). The results are representative of sections obtained from 5 tumors in the same group, scale bar: 100 μm (A and B). *P < 0.05 compared with control (C).
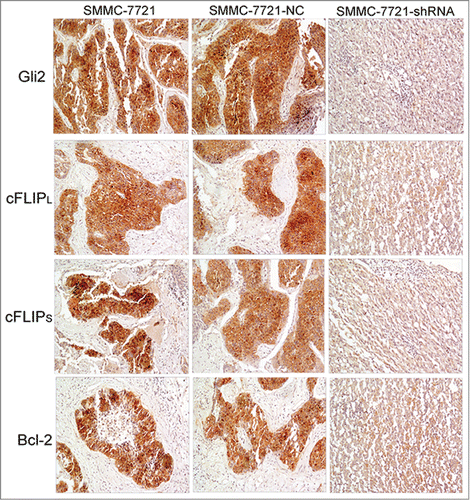
Association among c-FLIPL, c-FLIPS, Bcl-2, and Gli2 expressions in xenografts
Previous studies have demonstrated that Gli2 silencing enhanced TRAIL-induced apoptosis in SMMC-7721 cells and there was a significant downregulation of both mRNA and protein level of c-FLIP and Bcl-2 followed by Gli2 silencing in vitro. In this study, we further investigated whether Gli2 silencing affects the expression of c-FLIP and Bcl-2 in vivo. As shown in , Gli2 staining was much weaker in the SMMC-7721-shRNA group as compared with the SMMC-7721-NC and the SMMC-7721 groups. Likewise, stainings of c-FLIPL, c-FLIPS and Bcl-2 in the SMMC-7721-shRNA group were also weaker than those in the SMMC-7721-NC and the SMMC-7721 groups. Meanwhile, the tissue morphology of xenografts showed lower cell density and a smaller number of interstitial cells in the SMMC-7721-shRNA group, probablely because the increasing proportion of apoptotic cells, lower proliferative activity and slower growth rate of tumor.
Inhibition of Gli2 sensitizes SMMC-7721 cells to TRAIL-mediated apoptosis through down-regulation of c-FLIP and Bcl-2
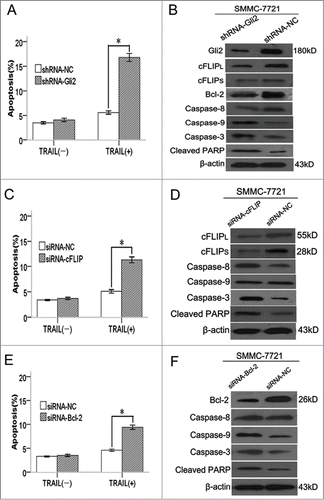
Flow cytometric analysis indicated that the apoptotic rate of shRNA-Gli2 cells with TRAIL treatment markedly increased to 16.81 ± 0.8% compared with 5.60 ± 0.3% in shRNA-NC cells (P < 0.05, ). Moreover, the expression levels of c-FLIPL, c-FLIPS, and Bcl-2 protein, which had been reported to confer resistance to TRAIL in some malignancies, were downregulated compared with those in shRNA-NC cells (). Because c-FLIPL, c-FLIPS, and Bcl-2 were down-regulated in SMMC-7721-shRNA-Gli2 cells, we analyzed the functional role of c-FLIP and Bcl-2 in shRNA-Gli2 facilitated TRAIL-mediated apoptosis. In the presence of TRAIL, SMMC-7721 cells transfected with c-FLIP-siRNA displayed much higer apoptotic rate (13.34 ± 0.5%) than cells transfected with NC-siRNA (5.12 ± 0.3%) (),as well as activation of caspase-8/-3 and cleaved PARP level (). Likewise, suppression of Bcl-2 by siRNA showed higer apoptotic rate in SMMC-7721 cells induced by TRAIL(9.45 ± 0.4% vs. 4.46 ± 0.2%). (), as well as activation of caspase-9/-3 and cleaved PARP level. (). These results suggest that inhibition of Gli2 sensitizes hepotoma cells to TRAIL-mediated apoptosis through downregulation of c-FLIP and Bcl-2.
Figure 5. Inhibition of Gli2 sensitizes SMMC-7721 cells to TRAIL-mediated apoptosis through down-regulation of c-FLIP and Bcl-2. (A, C, and E) TRAIL-induced apoptosis in SMMC-7721 cells transfected with shRNA-Gli2, siRNA-cFLIP and siRNA-Bcl-2, respectively. Apoptotic levels were determined 24 h after TRAIL (100 ng/ml) treatment. (B, D, and F) Western blot analysis of caspase-8, -9, and -3 and cleaved PARP protein. *P < 0.05 compared with control.
Gli2-dependent overexpression of c-FLIP or Bcl-2 attenuated TRAIL-induced apoptosis by suppression of caspase activity
To examine whether Gli2 can inhibite TRAIL-induced apoptosis in hepatoma cells, we employed pcDNA-Gli2 to stably express Gli2 in HepG2 cells. The results indicated that, in the presence of TRAIL, apoptosis rate was reduced from 16.23 ± 0.6% in HepG2 cells transfected with pcDNA3.1(+) to 5.67 ± 0.4% in HepG2 cells transfected with pcDNA-Gli2 (). The expression of Gli2, c-FLIPL, c-FLIPS, and Bcl-2 proteins were up-regulated in HepG2 cells transfected with pcDNA-Gli2 (). To further analyze whether c-FLIP and Bcl-2 affect the apoptosis of HCC cell downregulated by Gli2 expression, we established 2 plasmids of pcDNA-cFLIP and pcDNA-Bcl-2 for stable expression of c-FLIP and Bcl-2 in HepG2 cells. Stable over-expression of c-FLIP and Bcl-2 protein in HepG2 cells was detected by Western blot. Compared with HepG2 cells transfected with empty vector, cells stably expressing c-FLIP treated with TRAIL displayed a obvious decrease of apoptotic cell population from 16.42 ± 0.50% to 6.81 ± 0.35%, as well as cell stably expressing Bcl-2 (from 17.12 ± 0.70% to 5.43 ± 0.42%) ().
Figure 6. Gli2-dependent overexpression of c-FLIP or Bcl-2 attenuated TRAIL-induced apoptosis by suppression of caspase activity. (A, C, and E) Flow cytometry analysis of apoptosis. Triplicate experiments showed consistent results. (B, D, and F) Western blot analysis of caspase-8, -9, and -3 and cleaved PARP protein. β-actin was used as an internal control. *P < 0.05 compared with control.
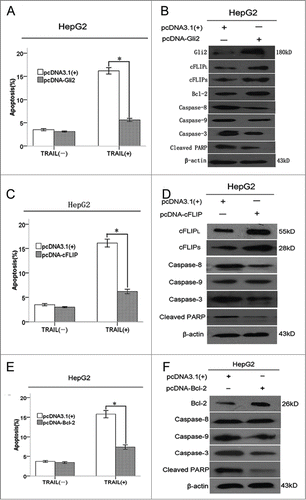
As c-FLIP and Bcl-2 have been reported to be a caspase substrate, we next investigated the correlation between caspase activity and over-expresion of c-FLIP or Bcl-2. Western blot demonstrated that overexpression of c-FLIP abrogated the activation of caspase-8, caspase-3, while over-expression of Bcl-2 attenuated the activation of caspase-9, caspase-3 (, upper panel). Furthermore, in the presence of TRAIL, lower levels of cleaved poly ADP-ribose polymerase (PARP) was found in HepG2 cells stably expressing c-FLIP or Bcl-2 (, lower panel). Collectively, these data suggest that Gli2-dependent overexpression of c-FLIP or Bcl-2 attenuated TRAIL-induced apoptosis by suppression of caspase activity.
Gli2 upregulated activity of c-FLIP gene promoter, -1007 to -244 was the important transcription regulatory region
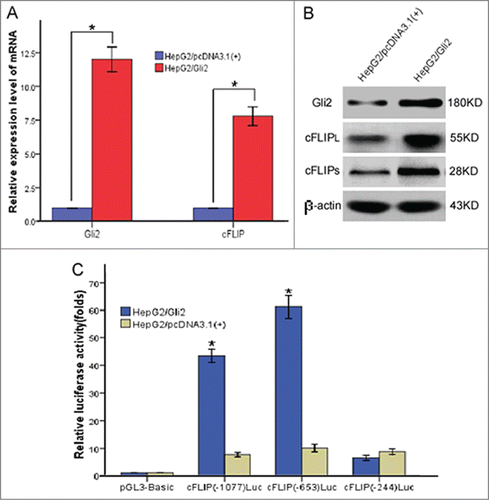
To verify the mechanism responsible for Gli2 up-regulated the expression of c-FLIP, we established stable expression of Gli2 (HepG2/Gli2) and empty vector (HepG2/pcDNA) in HepG2 cells by trasfection with pcDNA-Gli2 and pcDNA3.1(+), respectively. As shown in ,HepG2/Gli2 had increased levels of Gli2 expression when compared to cells that had been transfected with empty vector. Strinkingly, HepG2/Gli2 tremendously increased both mRNA and protein expression of c-FLIP, whereas the HepG2/pcDNA had no effect(). Luciferase reporter gene plasmids containing c-FLIP promoter, c-FLIP(-1077)Luc, c-FLIP(-653)Luc and c-FLIP(-244)Luc were used to transfection in HepG2/Gli2 and HepG2/pcDNA cells, respectively, with the pGL3-Basic vector as a control. The dual luciferase reporter gene system was used to analyze the effect of Gli2 on the activity of c-FLIP promoter. HepG2/Gli2 cells transfected with c-FLIP(-1077)Luc and c-FLIP(-653)Luc had significantly higher luciferase activity as compared with HepG2/pcDNA cells (, P < 0.05). However, c-FLIP(-244)Luc reporter plasmid was associated with decreased relative luciferase activity (, P < 0.05). These results suggest Gli2 significantly up-regulates the transcription activity of c-FLIP gene promoter, with a regulatory sequence between -1007 and -224 in the promoter region.
Figure 7. Gli2 upregulated activity of c-FLIP gene promoter, -1007 to -244 was the important transcription regulatory region. (A) Real time RT-PCR analysis of Gli2 and c-FLIP mRNA in stably transfected HepG2 cells with pcDNA 3.1(+)-Gli2 plasmids and control plasmids. 18s rRNA was used as an internal control. (B) Western blot analysis of Gli2, c-FLIPL,and c-FLIPS in stably transfected HepG2 cells with pcDNA 3.1(+)-Gli2 plasmids and control plasmids. β-actin was used as an internal control. (C) Luciferase reporter assays. Gli2 significantly up-regulated transcription activity of c-FLIP gene promoter, with a regulatory sequence by which Gli2 activated transcription between -1007 and -224 in the promoter region. Triplicate experiments showed consistent results.
Discussion
TRAIL is a new member of the TNF superfamily which has been discovered in recent years. TRAIL differs from TNF and Fas that it induces apoptosis only in virus-infected cells, transformed cells and tumor cells, and it shows almost no toxicity to normal tissue and cells.Citation4,19 This unique biological characteristic bring new hope to treatment of tumors, and it has the great potential to make a new generation of anti-tumor agents. Although TRAIL can strongly induce apoptosis in various types of tumor cells, almost all hepatoma cell lines are resistant to TRAIL to different extents. Therefore, TRAIL monotherapy, has extremely limited activity, and a combination therapy with TRAIL has to be found in the treatment of hepatoma.
Among all the interventional measures, combination chemotherapy is the most studied. The concept that chemotherapeutic agents enhance TRAIL-induced apoptosis in hepatoma cells has been recognized by many scientists. Shin et al. used TRAIL monotherapy to treat hepatoma cell lines, and found resistance to TRAIL in almost all the hepatoma cell lines. However, when cisplatin was combined, the resistance to TRAIL was altered. The underlying mechanism responsible for these phenomena is the chemotherapeutic agent activates the mitochondrial pathway and amplifys the TRAIL-mediated death receptor pathway, subsequently induces apoptosis of tumor cells.Citation20 Ganten et al. also reported that TRAIL had little effect on HepG2 and Hep3B cell lines without combination with 5-FU. Further investigation found that 5-FU alters the sensitivity to TRAIL in hepatoma.via up-regulation of DR5 and c-FLIP.Citation21 Other chemotherapeutic agent, such as adriamycin, camptothecin and paclitaxel were shown to be synergistic with TRAIL by Yamanaka et al. These authors postulated hepatoma cells were resistant to TRAIL at the death-inducing signaling complex (DISC) level, while the chemotherapeutic agents activated the activity of caspase-8 and increased the aggregation of DISC.Citation6 Further studies showed chemotherapeutic agents to up-regulate DR 5 expression in a non-p53-dependent manner and to inhibit activation of NF-κB, and thereby significantly enhanced the TRAIL-induced apoptotic effect.
Our early study on the activation status of the SHH pathway in hepatoma cell lines (HepG2, PLC/PRF/5 and SMCC-7721) found mRNAs and proteins of genes in this pathway. Shh, Ptch1, Smoh and Gli2, had higher expression levels in the PLC/PRF/5 and SMCC-7721 HCC cell lines compared to normal hepatic cells. Using shRNA to down-regulate the expression of transcription factor Gli2, the proliferation of HCC cells was inhibited, and the TRAIL-induced apoptosis enhanced. In addition, Gli2 was positively correlated with expressions of the apoptotic inhibitory genes c-FLIP and Bcl-2. To further confirm whether shRNA plasmids down-regulate Gli2 enhanced TRAIL-induced cell apoptosis in vivo, and to investigate the regulatory activity of Gli2 on the expression of c-FLIP, the present study demonstrated shRNA-mediated Gli2 down-regulation enhanced the anti-tumor effect of TRAIL, as reflected in the smaller volumes and lower weights of xenograf in the Gli2-shRNA group as compared to control group.
The results of immunohistochemistry (IHC) showed a significantly down-regulated Gli2 protein expression in the Gli2-shRNA group and a higher percentage of apoptotic cells, which were consistent with those seen in HE staining and TUNEL assay. Therefore, the enhancement of anti-tumor effect with TRAIL by downregulation of Gli2 might be related to induction of apoptosis.
Multiple studies have suggested over-expression of Bcl-2 led to resistance to TRAIL in HCC cells. The mechanism might be due to blockage of caspases-9, -7 and -3 activation, subsequently leading to inhibition of apoptosis.Citation22,23 The present study found a high expression level of Bcl-2 resulted in resistance to TRAIL and a decreased percentage of apoptotic cells in HepG2 cells. Also, a higher Bcl-2 expression led to inhibition of activation of caspases-9, and -3, and a lower level of the apoptotic marker cleaved PARP protein. In line with our data,Siegelin et al. showed ectopic overexpression of mouse Bcl-2 in protected glioma cells LN229 and U251 by KAAD-facilitated TRAIL-induced cytotoxicity and activation of caspase.Citation17 Down-regulation of Bcl-2 by Gli2 silencing has already been reported in hepatoma and prostate cancer.Citation24,25 In addition, Regl et al. reported the presence of a binding site to transcription factor Gli2 in the promoter region of Bcl-2, and that Gli2 activated Bcl-2 transcription.Citation26 Our IHC and Western blot results also showed Bcl-2 expression to alter with the up- or downregulation of Gli2 expression, thus providing indirect evidence that Bcl-2 was regulated by Gli2 expression.
C-FLIP, which has a highly similar structure and sequence to caspase-8, is another important regulatory factor in TRAIL-induced apoptosis. By competitive binding to FADD and/or caspase-8 with 2 DEDs in its N-terminal, c-FLIP prevented the formation of DISC and the activation of caspase-8, leading to termination of caspases cascade and inhibition of apoptosis indued by TRAIL.Citation27 Moreover, overexpression of c-FLIP decreased the sensitivity to TRAIL-induced apoptosis in some cancer cells, while down-regulation of c-FLIP reverse this phenomenon.Citation28-30 C-FLIP also be reported to mediate chemotherapeutic agent (5-FU and cyclopamine effect) in TAIL-induced apoptosis in HCC cells.Citation17,21 Our early cellular experiments also suggested Gli2 silencing enhanced TRAIL-induced apoptosis by down-regulating c-FLIP. Consistant with the finding above, IHC results confirmed that the expression level of c-FLIP protein reduced correspondingly in the Gli2-shRNA group. Moreover, cells became resistant to TRAIL, there was a lower percentage of apoptotic cells, caspases-8 and -3 activations were inhibited, and the apoptotic marker cleaved PARP protein was produced at a lower level, when HepG2 cells were transfected with c-FLIP. No previous studies have shown that Gli2 had a regulatory activity on the transcription of c-FLIP. Using the dual luciferase reporter gene system, we showed HepG2/Gli2 cells transfected with c-FLIP(-1077)Luc and c-FLIP(-653)Luc reporter plasmids had significantly higher relative luciferase activity than HepG2/pcDNA cells, while c-FLIP(-244)Luc reporter plasmid was associated with a reduced relative luciferase activity. This demonstrated Gli2 to significantly up-regulate the transcription activity of c-FLIP gene promoter, with a regulatory sequence by which Gli2 activated the transcription between -1007 to -244 in the promoter region.
In summary, shRNA-mediated silencing of Gli2 enhanced TRAIL-induced apoptosis in vivo through downregulation of c-FLIP and Bcl-2 expressions and Gli2 showed a regulatory activity on the transcription of c-FLIP. A regulatory site was found for Gli2 to be between -1007 to -244 of the promoter region.
Materials and Methods
Ethics statement
All animal work was conducted according to relevant national and international guidelines, including the requirements of the Association for the Assessment and Accreditation of Laboratory Animal Care International as described in the Guide for the Care and Use of Laboratory Animals, Eighth Edition. All animal protocols were approved by the internal Institutional Animal Care and Use Committee at the Second Affiliated Hospital of Guangzhou Medical College.
Cell culture and reagents
The human hepatoma cell lines HepG2 and SMMC-7721 were purchased from the Cell Bank of Shanghai Institute of Biochemistry &Cell Biology, the Chinese Academy of Sciences (Shanghai, China). The SMMC-772 cell line in which Gli2 was stably downregulated (SMMC-7721-shRNA) and the negative control cells (SMMC-7721-NC) were prepared and stored by our team. These cells were cultured in DMEM (Hyclone, USA), and supplemented with 10% fetal bovine serum (Hyclone, USA) at 37°C in a humidifed 5%CO2/95% air atmosphere. The recombinant human TRAIL (amino acids 114–281) was purchased from the PeproTech, Inc. (Rocky Hill, USA), and was reconstituted at 20 μg/ml in sterile PBS and stored at −20°C.
Establishment of the cell lines stably over-expressing Gli2, c-FLIP or Bcl-2 and siRNA transfections
A cDNA encoding human Gli2、c-FLIP or Bcl-2 was generated by PCR from human cDNA library and subcloned into the eukaryotic expression plasmid pcDNA3.1(+). The constructs were verified by DNA sequencing. HepG2 cells were stably transfected with pcDNA 3.1(+)-Gli2 plasmids, or control plasmid 3.1(+) vector using Lipofectamine2000 (Invitrogen, USA) following the manufacturer's specifications. Stable HepG2 cell lines over-expressing Gli2, c-FLIP or Bcl-2 was selected with changes of fresh medium containing 500 μg/mL G418 (Sigma, USA). Over-expression of Gli2, c-FLIP or Bcl-2 in the stable cell lines was examined by Western blot. SMMC-7721 cells were transfected with c-FLIP or Bcl-2 siRNA, or an equal amount of nonspecific control RNA as a control (Shanghai Biotech, Ltd. Corp., China). Transfection of siRNA was then performed using Lipofectamine2000 according to the manufacturer's protocol. After transfection with siRNA, cells were incubated for 24 h followed by incubation under the indicated conditions.
Tumor formation in nude mice
Five-week-old male BALB/c nude mices were purchased from the Slac Laboratory Animal (Shanghai, China). SMMC-7721-shRNA, SMMC-7721-NC, and SMMC-7721(1 × 106) were implanted subcutaneously into the right flank of mice. After 1 week, TRAIL (50 ng/mouse) was injected into the peritoneal space at days 1, 4, 8, 11, 15, and 18, respectively. Each group consisted of 5 mice, PBS treatment were used as control groups. Tumor size was monitored at intervals of 4 d. Tumor volume was determined by the formula: tumor length × tumor width2 × 0.5236 (tumor length, the tumor's longest diameter; tumor width, the shortest diameter perpendicular to tumor length). At the end, the mice were sacrificed by CO2 inhalation and tumors were resected, weighted, and subjected to hematoxylin staining, immunohistochemistry and tunel assay.
Detection of apoptosis in xenografts by TUNEL assay
The xenograft tissues were fixed with 10% formalin for 4 h and then embedded in paraffin. Hematoxylin-eosin (HE) staining was performed to estimate the pathological change and TUNEL was performed for detection of in situ apoptosis. The slices were deparaffinized in water and placed in 3% H2O2 for 5 min at room temperature. The TUNEL assay was carried out according to the manufacturer's protocol (Roche Molecular Biochemicals). At least 1000 cells from at least 10 scopes were counted using the DMR+Q550 system (Laica) technology. The apoptotic index was calculated. A positive result was brown or tan staining in the nucleus. The results from 3 independent experiments were averaged and statistically analyzed.
Immunohistochemistry
The tissue sections were dewaxed, rehydrated with xylene and graded alcohol, and then incubated in 3% hydrogen peroxide for 10 min to block endogenous peroxidase activity. Optimal antigen retrieval was carried out in citrate buffer (pH 6.0) for 10 min with a microwave oven to enhance immunoreactivity and then incubated in 10% blocking serum for 30 min at 37°C to reduce nonspecific binding. Anti-Gli2, anti-c-FLIP, anti-β-actin (Santa Cruz, USA) and anti-Bcl-2 (Cell Signaling Technology, USA) primary antibodies were incubated with the sections at 4°C overnight. Subsequently, biotinylated goat anti-rabbit immunoglobulins as secondary antibodies and streptavidin peroxidase complex reagent were applied. Finally, the visualization signal was developed with diaminobenzidine (DAB) and the slides were counterstained in hematoxylin. The Image-pro plus 6.0 system was used to analyze 4 fields randomly chosen from each slide. The images were amplified 100-fold, converted into gray-scale so as to distinguish the positive staining area from the background. The positive-stained area and the total area were measured by the system. The positive ratio was calculated as the staining area/total area × 100%. The stained area of each individual slide was determined by averaging the area ratio.
Flow cytometry analysis of apoptosis
HepG2 cells stably transfected with pcDNA 3.1(+)-Gli2, pcDNA 3.1(+)-c-FLIP or pcDNA 3.1(+)-Bcl-2 plasmids and corresponding control cells were seeded in the 6-wells and cultured for 24 h. SMMC-7721 cells were transfected with Gli2 shRNA, c-FLIP or Bcl-2 siRNA and incubated for 24 h. Then, the cells were administered with TRAIL (100 ng/ml) for another 24 h and collected. The cells were washed twice with ice cold PBS and resuspended in binding buffer. Annexin V-fluorescein isothiocyanate (FITC; 0.5 μg/ml) and propidium iodide (0.6 μg/ml) were then added to this cell suspension. After 15 min incubation in the dark at room temperature, stained cells were immediately analyzed by FACSCalibur (BD Biosciences, USA). All samples were assayed in triplicate.
RNA extraction and real time reverse transcription-polymerase chain reaction (Real time RT-PCR)
Total RNA was extracted using the TRIzol reagent (Invitrogen), the concentration and purity of the total RNA were detected with ultraviolet spectrophotometer. RT-PCR reaction was performed using a SYBR Premix Ex TaqTM kit (TaKaRa, Japan) according to the manufacturer's guidelines. First of all, 2 μg total RNA were obtained and underwent reserve transcription to compose cDNA. The PCR system (total amount 20 μL) included: Premix Ex TaqTM 10.0 μL, 0.8 μL of up-stream and down-stream primers each, added with double distilled water to 20 μL. The PCR reaction was catalyzed in lightcycler 480 real-time detection PCR analyzer at: 95°Cover 5 min for pre-denaturation; 92°C for 30s, and 60°C for 30s in a total of 40 cycles. Using 18s rRNA as an internal control gene, 2-ΔΔCt method was used to calculate relative expression levels of Gli2 or c-FLIP mRNA: ΔCt = Ct(Gli2)-Ct(18s rRNA), and ΔΔCt = HepG2/Gli2ΔCt-HepG2/pcDNA 3.1(+)ΔCt. The experimental results were means of triple independent repeated experiments. For the Gli2 gene, 4 μl of PCR product were run on a 2% agarose gel and visualized by ethidium bromide staining, the house-keeping gene GAPDH from each sample was used as an internal control. The primer sequences were: Gli2: sense 5′-TGGCCGCTTCAGATGACAGATGTTG-3′, antisense 5′- CGTTAGCCGAATGTCAGCCGTGAAG-3′; c-FLIP: sense 5′- GCACCCCAAAGTCAGAAAAA-3′, antisense 5′-ATGCTATCACCTCCCCTGTG-3′. 18s rRNA: sense 5′-CCTGGATACCGCAGCTAGGA-3′, antisense 5′- CACCCATTTGTGCATCACTC-3′. GAPDH: sense 5′- ATCTTCCAGGAGCGAGATCCC-3′, antisense 5′- CGTTCGGCTCAGGGATGACCT -3′. The primers were synthesized by the Shanghai Sangon Biological Engineering Technology & Services Co. Ltd.
Western blot
Antibodies against Gli2, c-FLIPL, c-FLIPS, and β-actin were purchased from the Santa Cruz Biotechnology, antibodies against Bcl-2, caspase8, caspase9 caspase3 and cleaved PARP were purchased from the Cell Signaling Technology,and the secondary antibodies conjugated to horseradish peroxidase were purchased from the Boster Biological Technology. Immunoblotting procedures were done as described previously.
Promoter constructs and luciferase reporter assays
For the construction of the c-FLIP reporter plasmid, 3 fragments of the putative promoter(-1077 to -48) were cloned into pGL3-Basic vector (Promega, USA). Each segment was approximately 410bp. KpnI and XhoI restriction sites were introduced to compose primer sequences. Upstream primers for 3 sequences to be amplified were: c-FLIP Promoter(-1077): 5′–ggggtacc TGAGTTGCAGCAGTCTGGAG -3′, c-FLIP Promoter(-653): 5′-ggggtacc ATCACTTGAGGCCAGCAGTT -3′, c-FLIP Promoter(-244): 5′-ggggtacc CGAGACCATTCTGGCCAAC -3′. Common downstream primer was: c-FLIP(-48): 5′– ccctcgag GACAGAGTCTCGCTCTGTGGC -3′. Products were 1030bp, 606bp and 197bp in size. Primers were composed by the Sangon Biotech (Shanghai) Co., Ltd. DNA-sequencing-confirmed the plasmids were named c-FLIP(-1077)Luc, c-FLIP(-653)Luc and c-FLIP(-244)Luc, which were stored for subsequent measurement with dual luciferase reporter gene assay. For luciferase reporter assays, each of 3 constructs and pGL3-Basic vector were transfected into HepG2/Gli2 and HepG2/pcDNA3.1(+) cells, respectively. Transfection was performed using Lipofectamine 2000 (Invitrogen, USA). We measured luciferase activity 48 h after transfection using the Luciferase Assay System (Promega, USA) according to the manufacturer's protocol and normalized it to the β-galactosidase expression level.
Statistical analysis
Data are expressed as mean ± SD and all statistical analyses were performed by using SPSS16.0. Comparisons among all groups were performed with one-way analysis of variance (ANOVA) test or unpaired Student's t-test. Differences were considered significant at P < 0.05. The results showed in each of the figures in this article are representative of at least the mean of 3 independent experiments.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the PhD Start-up Fund of Guangzhou Medical University, China (no. 2012C08), Medical Technology Project of Guangdong Province (no. B2014170) and Sun Yat-sen University Training Project, China (no. 13ykpy36).
References
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA: Cancer J Clin 2005; 55(2):74-108; PMID:15761078
- Page JM, Harrison SA. NASH and HCC. Clin Liver Dis 2009; 13(4):631-47; PMID:19818310
- Gores GJ, Kaufmann SH. Is TRAIL hepatotoxic? Hepatology 2001; 34(1):3-6; PMID:11431726
- Wang P, Song JH, Song DK, Zhang J, Hao C. Role of death receptor and mitochondrial pathways in conventional chemotherapy drug induction of apoptosis. Cell Signalling 2006; 18(9):1528-35; PMID:16442262
- Hao C, Song JH, Hsi B, Lewis J, Song DK, Petruk KC, Tyrrell DL, Kneteman NM. TRAIL inhibits tumor growth but is nontoxic to human hepatocytes in chimeric mice. Cancer Res 2004; 64(23):8502-6; PMID:15574753
- Yamanaka T, Shiraki K, Sugimoto K, Ito T, Fujikawa K, Ito M, Takase K, Moriyama M, Nakano T, Suzuki A. Chemotherapeutic agents augment TRAIL-induced apoptosis in human hepatocellular carcinoma cell lines. Hepatology 2000; 32(3):482-90; PMID:10960439
- Carlisi D, Lauricella M, D’Anneo A, Emanuele S, Angileri L, Di Fazio P, Santulli A, Vento R, Tesoriere G. The histone deacetylase inhibitor suberoylanilide hydroxamic acid sensitises human hepatocellular carcinoma cells to TRAIL-induced apoptosis by TRAIL-DISC activation. Eur J Cancer 2009; 45(13):2425-38; PMID:19643600
- Jin CY, Park C, Moon SK, Kim GY, Kwon TK, Lee SJ, Kim WJ, Choi YH. Genistein sensitizes human hepatocellular carcinoma cells to TRAIL-mediated apoptosis by enhancing Bid cleavage. Anti-Cancer Drugs 2009; 20(8):713-22; PMID:19617819
- Um HJ, Oh JH, Kim YN, Choi YH, Kim SH, Park JW, Kwon TK. The coffee diterpene kahweol sensitizes TRAIL-induced apoptosis in renal carcinoma Caki cells through down-regulation of Bcl-2 and c-FLIP. Chemico-Biol Interact 2010; 186(1):36-42; PMID:20403343; http://dx.doi.org/10.1016/j.cbi.2010.04.013
- Zhang S, Li G, Zhao Y, Liu G, Wang Y, Ma X, Li D, Wu Y, Lu J. Smac mimetic SM-164 potentiates APO2LTRAIL- and doxorubicin-mediated anticancer activity in human hepatocellular carcinoma cells. PloS One 2012; 7(12): e51461; PMID:23240027; http://dx.doi.org/10.1371/journal.pone.0051461
- Lu J, McEachern D, Sun H, Bai L, Peng Y, Qiu S, Miller R, Liao J, Yi H, Liu M, et al. Therapeutic potential and molecular mechanism of a novel, potent, nonpeptide, Smac mimetic SM-164 in combination with TRAIL for cancer treatment. Mol Cancer Ther 2011; 10(5):902-14; PMID:21372226; http://dx.doi.org/10.1158/1535-7163.MCT-10-0864
- Huang S, Okumura K, Sinicrope FA. BH3 mimetic obatoclax enhances TRAIL-mediated apoptosis in human pancreatic cancer cells. Clin Cancer Res: Off J Am Assoc Cancer Res 2009; 15(1):150-9; PMID:19118042; http://dx.doi.org/10.1158/1078-0432.CCR-08-1575
- Sicklick JK, Li YX, Jayaraman A, Kannangai R, Qi Y, Vivekanandan P, Ludlow JW, Owzar K, Chen W, Torbenson MS, et al. Dysregulation of the Hedgehog pathway in human hepatocarcinogenesis. Carcinogenesis 2006; 27(4):748-57; PMID:16339184
- Patil MA, Zhang J, Ho C, Cheung ST, Fan ST, Chen X. Hedgehog signaling in human hepatocellular carcinoma. Cancer Biol Ther 2006; 5(1):111-7; PMID:16397407
- Huang S, He J, Zhang X, Bian Y, Yang L, Xie G, Zhang K, Tang W, Stelter AA, Wang Q, et al. Activation of the hedgehog pathway in human hepatocellular carcinomas. Carcinogenesis 2006; 27(7):1334-40; PMID:16501253
- Ikram MS, Neill GW, Regl G, Eichberger T, Frischauf AM, Aberger F, Quinn A, Philpott M. GLI2 is expressed in normal human epidermis and BCC and induces GLI1 expression by binding to its promoter. J Invest Dermatol 2004; 122(6):1503-9; PMID:15175043; http://dx.doi.org/10.1111/j.0022-202X.2004.22612.x
- Siegelin MD, Siegelin Y, Habel A, Rami A, Gaiser T. KAAD-cyclopamine augmented TRAIL-mediated apoptosis in malignant glioma cells by modulating the intrinsic and extrinsic apoptotic pathway. Neurobiol Dis 2009; 34(2):259-66; PMID:19385057; http://dx.doi.org/10.1016/j.nbd.2009.01.012
- Zhang D, Liu J, Wang Y, Chen J, Chen T. shRNA-mediated silencing of Gli2 gene inhibits proliferation and sensitizes human hepatocellular carcinoma cells towards TRAIL-induced apoptosis. J Cell Biochem 2011; 112(11):3140-50; PMID:21695716; http://dx.doi.org/10.1002/jcb.23240
- Ashkenazi A, Holland P, Eckhardt SG. Ligand-based targeting of apoptosis in cancer: the potential of recombinant human apoptosis ligand 2Tumor necrosis factor-related apoptosis-inducing ligand (rhApo2LTRAIL). J Clin Oncol: Off J Am Soc Clin Oncol 2008; 26(21):3621-30; PMID:18640940; http://dx.doi.org/10.1200/JCO.2007.15.7198
- Shin EC, Seong YR, Kim CH, Kim H, Ahn YS, Kim K, Kim SJ, Hong SS, Park JH. Human hepatocellular carcinoma cells resist to TRAIL-induced apoptosis, and the resistance is abolished by cisplatin. Exp Mol Med 2002; 34(2):114-22; PMID:12085986; http://dx.doi.org/10.1038/emm.2002.17
- Ganten TM, Haas TL, Sykora J, Stahl H, Sprick MR, Fas SC, Krueger A, Weigand MA, Grosse-Wilde A, Stremmel W, et al. Enhanced caspase-8 recruitment to and activation at the DISC is critical for sensitisation of human hepatocellular carcinoma cells to TRAIL-induced apoptosis by chemotherapeutic drugs. Cell Death Differ 2004; 11 Suppl 1:86-96; PMID:15105837; http://dx.doi.org/10.1038/sj.cdd.4401437
- Fulda S, Meyer E, Debatin KM. Inhibition of TRAIL-induced apoptosis by Bcl-2 overexpression. Oncogene 2002; 21(15):2283-94; PMID:11948412; http://dx.doi.org/10.1038/sj.onc.1205258
- Han SI, Kim YS, Kim TH. Role of apoptotic and necrotic cell death under physiologic conditions. BMB Rep 2008; 41(1):1-10; PMID:18304444; http://dx.doi.org/10.5483/BMBRep.2008.41.1.001
- Kim Y, Yoon JW, Xiao X, Dean NM, Monia BP, Marcusson EG. Selective down-regulation of glioma-associated oncogene 2 inhibits the proliferation of hepatocellular carcinoma cells. Cancer Res 2007; 67(8):3583-93; PMID:17440069; http://dx.doi.org/10.1158/0008-5472.CAN-06-3040
- Narita S, So A, Ettinger S, Hayashi N, Muramaki M, Fazli L, Kim Y, Gleave ME. GLI2 knockdown using an antisense oligonucleotide induces apoptosis and chemosensitizes cells to paclitaxel in androgen-independent prostate cancer. Clin Cancer Res: Off J Am Assoc Cancer Res 2008; 14(18):5769-77; PMID:18794086; http://dx.doi.org/10.1158/1078-0432.CCR-07-4282
- Regl G, Kasper M, Schnidar H, Eichberger T, Neill GW, Philpott MP, Esterbauer H, Hauser-Kronberger C, Frischauf AM, Aberger F. Activation of the BCL2 promoter in response to HedgehogGLI signal transduction is predominantly mediated by GLI2. Cancer Res 2004; 64(21):7724-31; PMID:15520176; http://dx.doi.org/10.1158/0008-5472.CAN-04-1085
- Kim Y, Suh N, Sporn M, Reed JC. An inducible pathway for degradation of FLIP protein sensitizes tumor cells to TRAIL-induced apoptosis. J Biol Chem 2002; 277(25):22320-9; PMID:11940602; http://dx.doi.org/10.1074/jbc.M202458200
- Roth W, Reed JC FLIP protein and TRAIL-induced apoptosis. Vitam Horm 2004; 67:189-206.
- Zhang X, Jin TG, Yang H, DeWolf WC, Khosravi-Far R, Olumi AF. Persistent c-FLIP(L) expression is necessary and sufficient to maintain resistance to tumor necrosis factor-related apoptosis-inducing ligand-mediated apoptosis in prostate cancer. Cancer Res 2004; 64(19):7086-91; PMID:15466204; http://dx.doi.org/10.1158/0008-5472.CAN-04-1498
- Brooks AD, Sayers TJ. Reduction of the antiapoptotic protein cFLIP enhances the susceptibility of human renal cancer cells to TRAIL apoptosis. Cancer Immunol Immunother 2005; 54(5):499-505; PMID:15614529; http://dx.doi.org/10.1007/s00262-004-0595-8