Abstract
Somatic cells senesce in culture after a finite number of divisions indefinitely arresting their proliferation. DNA damage and senescence increase the cellular number of centrosomes, the 2 microtubule organizing centers that ensure bipolar mitotic spindles. Centrosomes also provide the basal body from which primary cilia extend to sense and transduce various extracellular signals, notably Hedgehog. Primary cilium formation is facilitated by cellular quiescence a temporary cell cycle exit, but the impact of senescence on cilia is unknown. We found that senescent human fibroblasts have increased frequency and length of primary cilia. Levels of the negative ciliary regulator CP110 were reduced in senescent cells, as were levels of key elements of the Hedgehog pathway. Hedgehog inhibition reduced proliferation in young cells with increased cilium length accompanying cell cycle arrest suggesting a regulatory function for Hedgehog in primary ciliation. Depletion of CP110 in young cell populations increased ciliation frequencies and reduced cell proliferation. These data suggest that primary cilia are potentially novel determinants of the reduced cellular proliferation that initiates senescence.
Abbreviations
| CP110 | = | centriolar coiled coil protein of 110kDa |
| DABCO | = | 1,4-Diazabicyclo[2.2.2]octane |
| DAPI | = | 4′,6-diamidino-2-phenylindole |
| ECL | = | enhanced chemiluminescence |
| FITC | = | Fluorescein isothiocyanate |
| GAPDH | = | glyceraldehyde 3-phosphate dehydrogenase |
| Hh | = | Hedgehog |
| HMEC | = | human mammary epithelial cell |
| NHDF | = | normal human dermal fibroblasts |
| PLK4 | = | Polo-like kinase 4 |
| SA-β-gal | = | senescence-associated β-galactosidase |
| SAHF | = | senescence-associated heterochromatin foci |
| Smo | = | smoothened |
Introduction
The finite replicative lifespan of somatic cells is a long-established phenomenon.Citation1 After an extended culture period, cells undergo a stable and indefinite exit from the cell cycle and manifest additional physiological alterations that indicate senescence.Citation2,3 Senescence can arise through persistent DNA damage signaling, notably at telomeric sequences that can become exposed after multiple divisionsCitation4-6 or through a response to strong mitogenic signaling induced by oncogenes.Citation7,8 Senescence-induced changes include the increased expression of senescence-associated ß-galactosidase (SA-β-gal);Citation9 increased levels of the tumor suppressor p16INK4a Citation10,11 and the formation of senescence-associated heterochromatin foci (SAHF).Citation12
An additional impact of cellular senescence is an increase in the number of centrosomes per cell, an effect that is also seen when cells suffer DNA damage through a range of genotoxic insults.Citation13,14 Mitotic centrosomes contain 2 centrioles embedded in a pericentriolar matrix. Centrioles are barrel-shaped assemblies of 9 microtubule triplets that taper to doublets at their distal end. After cell division, the individual centrioles dissociate to serve as templates on which new centrioles will form during the next S phase.Citation15 This means that each centrosome will contain a mature mother centriole and an immature daughter. Primary cilia are microtubule-based organelles that extend from the surface of most human cell types to sense and transduce various extracellular signals.Citation16,17 They arise from a template provided by the mature centriole docked to the plasma membrane, the basal body.Citation16-18 The cilium core, the axoneme, consists of 9 microtubule doublets that extend from the basal body. The axoneme is bounded by membrane and a diffusion barrier maintains a cilium-specific distribution of proteins within this membrane subregion.Citation19 In particular, signaling through the development-regulating Hedgehog (Hh) pathway requires the controlled access of the Hh effector, Smoothened (SMO), to the primary cilium.Citation17,Citation20-23
Cilium formation is closely regulated and linked to the cell cycle, as cilia must be resorbed to allow the mitotic functioning of centrosomes in bipolar spindle formation. Cellular quiescence, a temporary exit from the cell cycle that can be induced by the removal of growth factors, facilitates ciliogenesis.Citation18,24 Here, we explore the impact on cilia of the more permanent cell cycle arrest caused by replicative senescence in primary human fibroblasts.
Results
To obtain cells that had undergone replicative senescence, we monitored the doubling times of 3 primary human fibroblast lines, BJ, MRC5 and NHDF, over extended culture periods. As shown in , their doubling times eventually increased to an extent where the bulk of the population was no longer proliferating. We confirmed that this was due to the induction of cellular senescence by testing for senescence-associated heterochromatin foci and β-galactosidase activity (Fig. S1A and B), which serve as cellular markers of senescence.Citation2 Based on these analyses, in the rest of the paper we define the following as ‘senescent’: BJ: ≥250 d in culture; MRC5: ≥100 d; NHDF: ≥80 d. ‘Proliferating’ cells were BJ: < 120 d in culture; MRC5: < 30 d; NHDF: < 20 d. We observed elevated percentages of γ-H2AX foci in senescent BJ cells, consistent with the DNA damage response that accompanies cellular senescence,Citation4,6 with an increased number of centrioles in those cells with γ-H2AX foci (). Senescence-associated centrosome abnormalities associated with aneuploidy have been reported in other human fibroblast lines,Citation14 so that this centrosomal amplification is a likely consequence of elevated DNA damage signaling in senescing human cells.Citation13
Figure 1. Increased frequency and length of cilia in senescent human fibroblasts; (A) Doubling time over extended culture periods of BJ, MRC5 and NHDF cells; (B). Immunofluorescence microscopy of senescent BJ cells stained with antibodies to Cep135 (green) and γ-H2AX (red). DNA was visualised with DAPI (blue). Scale bar, 10 μm; (C) Quantitation of the frequency of centrosomal amplification in senescent BJ cells with γ-H2AX staining, scored as >4 centrin2 spots in a cell. Histograms show means ± s.d. of 3 separate experiments in which at least 200 cells were quantitated; (D) Quantitation of the ciliation frequency in the indicated cells, based on imaging of acetylated tubulin. ‘Unt’, untreated. Serum starvation (‘Ser.-stvd’) consisted of 24 h culture with 0.1% newborn calf serum. Histograms show means ± s.d. of 3 separate experiments in which at least 200 cells were quantitated; (E) Immunofluorescence microscopy of the indicated cells stained with antibodies to Cep135 (green) and acetylated tubulin (red). DNA was visualised with DAPI (blue). Scale bar, 10 μm; (F) Quantitation of cilium length in the indicated cell lines. At least 30 ciliated cells were scored for each. *, P < 0.05; **, P < 0.01; ***, P < 0.001 compared with controls of the same cell line by unpaired t-test.
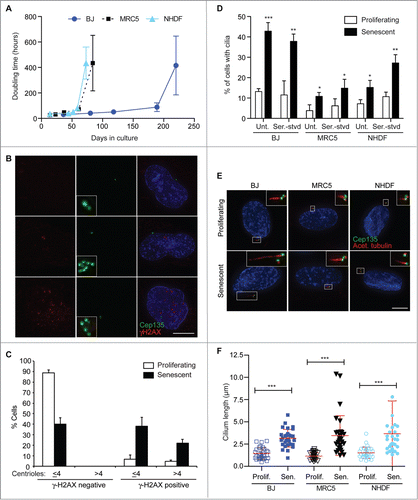
Given the close relationship between centrioles and basal bodies, we next asked if primary cilia were altered in senescent cells. The frequency with which primary cilia arose was significantly increased in senescent populations of BJ, MRC5 and NHDF cells, with little additional ciliation occurring when the populations were induced to quiesce by 24h serum starvation (). Longer starvation periods led to higher levels of ciliation in proliferating cells, in keeping with previously published data on cilium induction through cellular quiescence (Fig. S1C).Citation18,24 When we analyzed these cilia by immunofluorescence microscopy, we found that they were consistently longer in senescent cells, with the overall mean length being 3.2 ± 1.6 μm in senescent cells as against 1.4 ± 0.6 μm in proliferating controls (). Immunofluorescence microscopy indicated otherwise normal centriolar/ basal body and ciliary composition (Fig. S1D).
The presence of a cilium on a given cell was not associated with the presence of γ-H2AX foci (), suggesting that persistent DNA damage response signaling is not a prerequisite for ciliation. However, extra centrioles, such as those induced by DNA damage or overexpression of the centriole-regulatory polo-like kinase 4 (PLK4), can lead to multiple cilia being formed in a single cell.Citation25,26 We found that 24.8 ± 2.7% of senescent BJ cells had multiple cilia (; N = 3 experiments in which 200 cells were analyzed). No such structures were seen in the control analysis of proliferating BJ cells. Together, these data demonstrate a markedly increased level of primary cilium surface area in senescent human fibroblasts.
Figure 2. Cell-intrinsic control of extended cilium length in senescent cells; (A) Immunofluorescence microscopy of senescent BJ cells stained with antibodies to Arl13b (green) and γ-H2AX (red). DNA was visualised with DAPI (blue). Scale bar, 10 μm; (B) Quantitation of ciliation frequency in senescent BJ cells with γ-H2AX staining. Histograms show means ± s.d. of 3 separate experiments in which at least 200 cells were quantitated; (C) Immunofluorescence microscopy of senescent BJ cells stained with antibodies to centrin3 (red) and Arl13b (green) or Cep135 (red) and acetylated tubulin (green), as indicated. DNA was visualised with DAPI (blue). Scale bar, 10 μm; (D) Quantitation of cilium length in BJ cells, before or 24 h after the washout of 72 h 4 mM chloral hydrate treatment. This chloral hydrate treatment caused the removal of 90% of cilia from both proliferating and senescent cells. At least 30 ciliated cells were scored for each condition. **, P < 0.01; ***, P < 0.001 in comparison to the indicated controls by unpaired t-test; (E) Immunoblot analysis of CP110 expression in BJ cells. Size markers are indicated in kDa.
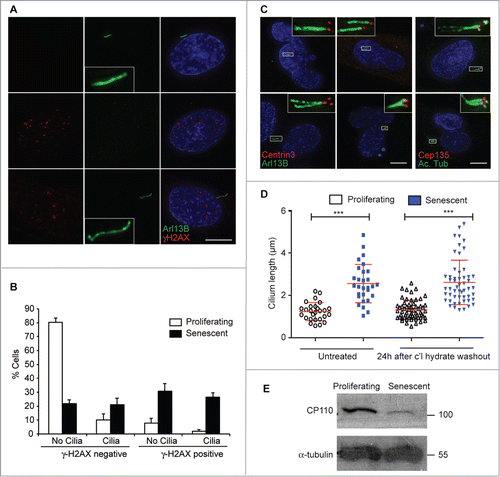
The role of the centrosome in formation of the bipolar spindle necessitates that the cilium be lost during mitosis, so that a more limited time for ciliary elongation is available to proliferating cells than to cells which are no longer cycling. To understand how senescent cells acquire their elongated cilia, we tested if an extended period without cycling was required for such ciliation. We used chloral hydrate to deciliate proliferating and senescent BJ fibroblasts.Citation27 72 h after chloral hydrate treatment, the ciliation frequency was reduced from 17 ± 5% to 2 ± 1% in proliferating cells and from 44 ± 6% to 1 ± 1% in senescent cells (N = 3). We then washed out the drug and allowed cells to acquire cilia again. Cilia were restored to equivalent percentages of cells and notably, cilia lengths were re-established at the previously-existing length at 24h after drug washout (). As this is shorter than the cell cycle period in BJ cells, this observation suggests a cell-intrinsic cilium length setting that is independent of the time spent outside cycle. As a candidate for controlling this length setting, we tested whether the levels of CP110, a negative regulator of ciliogenesis Citation28,29 that also localizes to centrosomes,Citation30 were altered. Quantitative RT-PCR analysis revealed that senescent cells expressed 0.26 ± 0.10 (N = 3) of the levels of CP110 that were expressed in proliferating controls and, as shown in , CP110 protein levels were greatly reduced in aged cells.
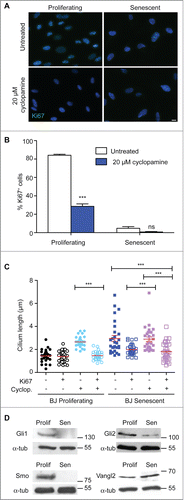
The primary cilium is established as the key organelle at which Hh signaling takes place.Citation21,31 Given the pro-mitogenic role established for Hh,Citation32,33 we therefore tested whether inhibition of Hh through the small molecule inhibitor, cyclopamine,Citation34 affected fibroblast proliferation, as determined by Ki67 staining. As shown in , cyclopamine caused a decline in the proliferative index in both young and senescent BJ populations. The few Ki67+ cells in the senescent populations had shorter cilia than those no longer in cycle (). Notably, the mean cilium length in those young fibroblasts that were no longer proliferating after cyclopamine treatment became as long as it was in senescent cells, while cilia in untreated cells were the same length in Ki67+ and Ki67− cells (). Strikingly, when we explored if any other ciliary regulators were differentially expressed as cells became senescent by testing a panel of 84 cilium-specific genes by quantitative RT-PCR (Fig. S2), the most significant downregulation occurred in the expression of the GLI2 gene, which encodes a downstream component of the Hh signaling pathway. Immunoblot analysis of GLI1, GLI2 and SMO expression confirmed the general loss of Hh components in senescent cells (), with the increase in expression of Vangl2 protein confirming our RT-PCR analysis with respect to the Wnt signaling pathway. Together, these data suggest that the loss of Hh signaling leads to reduced proliferation in human fibroblasts. The increased ciliary surface area provided by both the extended length and the higher number of cilia in senescent cells may impede mitogenic Hh signaling by diluting out components of the receptor pathway.Citation26
Figure 3. Hedgehog inhibition blocks proliferation and increases cilium length; (A) Immunofluorescence microscopy of BJ cells stained with antibodies to Ki67 (green). DNA was visualised with DAPI (blue). Scale bar, 10 μm; (B) Quantitation of the proliferative index of BJ cells after the indicated treatment, as determined by microscopy analysis of Ki67 signal. Histograms show means ± s.d. of 3 separate experiments in which at least 100 cells were quantitated; (C) Quantitation of cilium length in the indicated cells. At least 30 ciliated cells were scored for each condition, even when there were very few such cells. ***, P < 0.001 in comparison to untreated or indicated controls by unpaired t-test. ‘ns’, not significant; (D) Immunoblot analysis of the indicated protein expression in BJ cells. Size markers are indicated in kDa.
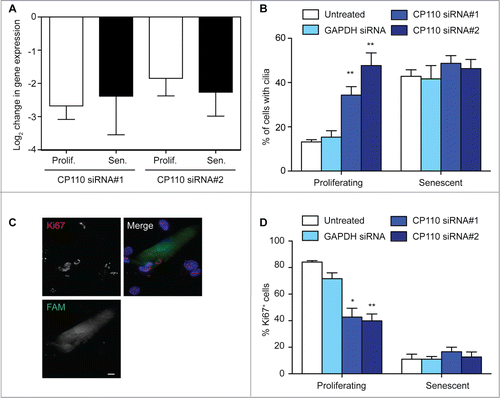
With this idea in mind, we used siRNA to deplete CP110 from BJ fibroblasts to test the impact of increased ciliation on proliferation (). Knockdown of CP110 caused an increased level of ciliation in proliferating, but not in senescent populations (). We also observed small but statistically significant increases in the mean cilium length after CP110 depletion, from 1.2 ± 0.06 μm (N = 37; GAPDH siRNA) to 1.6 ± 0.07 μm (N = 30; CP110 siRNA #1; P < 0.001) and 1.4 ± 0.09 μm (N = 30; CP110 siRNA #2; P < 0.05). Strikingly, CP110 knockdown caused a marked decline in the number of proliferating cells (). These data suggest that increased ciliation, resulting from the loss of CP110, leads to a decline in proliferative index that may potentiate cellular senescence. Overexpression of CP110 caused a decline in the fraction of ciliated cells, but did not affect either mean cilium length or the number of proliferating cells in either young or old populations (Fig. S3), indicating that senescence, once established, cannot simply be reversed through cilium manipulation.
Figure 4. CP110 depletion causes increased ciliation and reduced cellular proliferation; (A) Quantitation of CP110 depletion in BJ cells by siRNA as determined by quantitative RT-PCR. Knockdowns are compared to the effect of GAPDH siRNA normalized to housekeeping gene expression. Data show the mean—s.d. of 3 separate experiments; (B) Quantitation of the ciliation frequency in BJ cells after the indicated treatments. Histograms show means + s.d. of 3 separate experiments in which at least 100 cells were quantitated; (C) Microscopy of CP110 siRNA-transfected cells stained with antibodies to Ki67 (red in merge). Fluorescent RNA (FAM; green) was cotransfected at a ratio of 1:5 with the siRNA as a transfection control. DNA was visualised with DAPI (blue). Scale bar, 10 μm; (D) Quantitation of the proliferative index of BJ cells after the indicated treatment, as determined by microscopy analysis of Ki67 signal. Histograms show means ± s.d. of 3 separate experiments in which at least 100 transfected cells were quantitated; *, P < 0.05; **, P < 0.01 in comparison with GAPDH siRNA controls by unpaired t-test.
Discussion
The centrosomal abnormalities seen here and in previous work that analyzed senescent human fibroblastsCitation14 are similar to those seen in cells that have been subjected to genotoxic stress.Citation35 As senescent populations also show DNA damage responses,Citation4,8 a DNA damage-induced cell cycle arrest permissive for centrosome reduplication as an early event in senescence induction is consistent with these observations. Primary cilia arise while cells are no longer cycling and the frequent aberrant cilia seen in senescent cells may be attributable, at least in part, to these centrosomal abnormalities, as we have seen previously in irradiation experiments.Citation25
Our data indicate that ciliation is associated with cellular senescence in human fibroblasts and that expression of Hh signaling pathway components is reduced in senescent populations. Hh signaling has been ascribed a mitogenic role through activated GLI2 blockade of the growth-inhibitory p16INK4a promoter.Citation32 The involvement of Hh in a range of cell renewal activities throughout the lifespan of an organism has led to its being invoked as an antagonist of aging.Citation36 While our data indicating the involvement of Hh signaling in continued proliferation are consistent with this model, we did not observe the loss of primary cilia in senescent cells that was described in human mammary epithelial cell (HMEC) cultures.Citation32 Senescing HMECs and fibroblasts have distinct programmes of gene expression, so the impact of ciliary signaling may differ depending on the cell type.Citation37 Although some primary cilia, such as those carried by HMECs and embryonic stem cells, drive pro-mitogenic Hh signaling,Citation38 current evidence suggests that ciliation may be associated more generally with reduced proliferation.Citation39 Tumor cells frequently lack cilia.Citation40 The mitotic kinase Aurora A causes disassembly of the primary cilium Citation41 and cilium-dependent cell cycle exit through Aurora A inhibition has been described as a means of inducing quiescence in human cells.Citation42 Clearly, in the senescent cells we analyzed here, the Hh pathway is markedly downregulated, despite the presence of primary cilia.
While the acquisition of a primary cilium may be a transient event in a proliferating cell population,Citation24 extending cilium length may be a novel mechanism to establish senescence. If a threshold length of cilium is still responsive to a mitogen such as Hh, a lengthening of the structure may contribute to an effective dilution of this signaling capacity Citation26 and a blunting of the signal. A cooperative model in which this downregulation of Hh inhibits transcriptional activation of Gli to further inhibit mitogenic Hh signaling, may establish a permanent cell cycle exit. Such a model implicates the regulation of primary ciliation in the control of cellular senescence.
Materials and Methods
Cloning
For CP110 cDNA cloning, RNA was extracted from BJ cells using TRIzol reagent (Invitrogen). Reverse transcription was performed using High Capacity RNA to cDNA Kit (Applied Biosystems) and PCR with KOD Hot Start polymerase. The cDNA was cloned into pEGFP-C1 (Clontech) and verified by sequencing. The primers used to amplify cDNA were: CP110; 5′ CCGTCGACATGGAGGAGTATGAGAAG 3′ and 5′ CCGGATCCAATTGTCGCAACATTGG 3′]. For transient transfection, 2 μg of plasmid DNA were complexed with lipofectamine in serum-free Optimem (Invitrogen) and added to cells at 70–80% confluency. Fresh media was added 6 h after transfection. Cells were analyzed 48 h after transfection.
Cells and cell culture
Primary human BJ, MRC5 and NHDF fibroblasts were obtained from ATCC. Cells were grown in DMEM medium supplemented with 10% fetal bovine serum (Lonza) and 1% penicillin/streptomycin (Sigma-Aldrich). Cells were grown in a 5% CO2, 37°C incubator. To deplete cells of serum, cells were washed 3 times in warm PBS before adding DMEM medium with 0.1% newborn calf serum (NCS). SA-β-gal positive cells were stained as described.Citation9 Cyclopamine (Santa Cruz) was dissolved in 100% ethanol and used at 20 μM for 48 h. Chloral hydrate (Fisher Scientific) was dissolved in Millipore water and used at 4 mM for 72 h, with fresh chloral hydrate being replaced every 12 h. For washout experiments, cells were washed 3 times in warm PBS before adding fresh DMEM medium for 24 h.
Immunoblotting
Whole-cell extracts were prepared with radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 7.4, 1% NP-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, and protease inhibitor cocktail). Extracts were boiled in loading buffer for 5 mins. Proteins were separated on 7.5% SDS PAGE gels and transferred to nitrocellulose membranes (GE Healthcare) for analysis. Blots were detected by ECL (GE Healthcare). Monoclonal primary antibodies used were: mouse anti- α-tubulin (B512, Sigma-Aldrich, 1:10000) and rabbit anti-CP110 (Q-12, Santa Cruz, 1:100).
RNA-mediated interference
BJ cells were co-transfected with Silencer Select siRNAs (Ambion) inhibitory to CP110; #1 Sense 5′ GCAAAACCAGAAUACGAGATT 3′; Antisense 5′ UCUCGUAUUCUGGUUUUGCAT 3′ and #2 Sense 5′ CAAGCGGACUCACUCCAUATT 3′; Antisense 5′ UAUGGAGUGAGUCCGCUUGAG 3′] or GAPDH as an experimental control; Sense 5′ UGGUUUACAUGUUCCAAUATT 3′; Antisense 5′ UAUUGGAACAUGUAAACCATG 3′] and a FAM (carboxyfluorescein) labeled transfection control using lipofectamine (Invitrogen). 50 nmol of siRNA were complexed with lipofectamine in serum-free Optimem (Invitrogen) and added to cells at 70–80% confluency. Fresh media was added 6 h after transfection. Cells were analyzed by microscopy or qPCR 48 h after transfection.
Microscopy
BJ cells were grown on glass coverslips for 24 h prior to fixation in methanol/5 mM EGTA at −20°C for 10 min. Alternatively, cells were fixed in 4% PFA for 10 min and permeabilized in 0.15% Triton X-100 in PBS for 2 min. Prior to fixation and staining with acetylated tubulin, cells were incubated on ice for 30 min to depolymerize microtubules. The cells were blocked in 1% BSA in PBS and incubated with primary antibodies for 1 h at 37°C followed by a 45-min incubation at 37°C with secondary antibodies. Secondary antibodies were labeled with FITC and Alexa 594 (Jackson ImmunoResearch Laboratories, Inc.). Slides were mounted with DABCO (2.5% DABCO (Sigma-Aldrich), 50 mM Tris-HCl, pH 8, and 90% glycerol) and supplemented with 1 μg/ml DAPI. Cells were imaged on an integrated microscope system (DeltaVision) controlled by SoftWorx software (Applied Precision) mounted on a microscope (IX71; Olympus) with a PlanApo N100× oil objective (NA 1.40). Images were taken using a camera (CoolSnap HQ2; Photometrics), deconvolved in SoftWorx using the ratio method and maximal intensity projections were converted to TIFFs using Volocity (Perkin Elmer). Cell counting was performed on an Olympus BX51 microscope using a 100× oil (NA 1.35) objective. Cilium length was manually measured from the tip to the basal body using the “Line Tool” in Volocity. Monoclonal antibodies used were as follows: Centrin3 (3E6, Abnova, 1:1000), acetylated tubulin (T6793, Sigma-Aldrich, 1:2000), γ-H2AX (JBW301; Millipore, 1:1000) and Ki67 (SP6, Abcam, 1:50). Polyclonal antibodies used were against γ-tubulin (T3559; Sigma, 1:1000), centrin2 (N-17, Santa Cruz, 1:500), Cep135 (ab75005, Abcam, 1:1000), Arl13b (17711–1-AP, Proteintech, 1:500), IFT88 (13967–1-AP, 1:800, Proteintech), Pericentrin (ab4448, Abcam, 1:1000), GFP (ab6556, Abcam, 1:500) and Tri-MethyL-Histone H3 Lys9 (#9754, Cell Signaling, 1:800).
Real-Time PCR
RNA was extracted by direct addition of lysis buffer to cultured cells using an RNeasy mini kit (Qiagen, Crawley, UK). cDNA was generated using a Qiagen/SABiosciences RT2 First Strand Kit in a 20 μl reaction using 1 μg of the total RNA. One μl of the cDNA synthesis reaction was added to 12.5 μl RT2 SYBR Green Mastermix with 1 μl of the 10 μM primer pair in a 25 μl reaction. Primers for CP110 were obtained from Primerdesign (Southampton, UK): Sense Primer; 5′ GGACCAAGTGCTCTCAAAGG 3′]; Antisense primer; 5′ TCTGAAAGCTGCCGTTTAGTT 3′]. The amplification conditions used were 15 s at 95°C and 1 min at 60°C for 40 cycles. Relative gene expression was analyzed using the 2−ΔΔCT method. CT values for housekeeping genes and for the genes of interest were measured in proliferating and senescent samples. The CT values for each gene of interest were normalized to the housekeeping genes (ΔCT) and then compared to that of the control proliferating sample (ΔΔCT). The fold change in gene expression was calculated using 2(−ΔΔCT). Where the fold change is greater than 1, the result represents a fold up-regulation in gene expression. Where the fold change is less than 1, the negative inverse of the result represents a fold down-regulation. The standard deviations were also calculated in the same way.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental_Files
Download Zip (3.9 MB)Acknowledgments
We thank Emmanuelle Passegué and Donncha Dunican for comments on the manuscript.
Supplemental Materials
Supplemental materials may be found here: http://dx.doi.org/10.4161/15384101.2015.945868
Additional information
Funding
References
- Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp Cell Res 1965; 37:614-36; PMID: 14315085; http://dx.doi.org/10.1016/0014-4827(65)90211-9
- Rodier F, Campisi J. Four faces of cellular senescence. J Cell Biol 2011; 192:547-56; PMID: 21321098; http://dx.doi.org/10.1083/jcb.201009094
- Campisi J, d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol 2007; 8:729-40; PMID: 17667954; http://dx.doi.org/10.1038/nrm2233
- d'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature 2003; 426:194-8; PMID: 14608368; http://dx.doi.org/10.1038/nature02118
- Herbig U, Jobling WA, Chen BP, Chen DJ, Sedivy JM. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a). Mol cell 2004; 14:501-13; PMID: 15149599; http://dx.doi.org/10.1016/S1097-2765(04)00256-4
- Fumagalli M, Rossiello F, Clerici M, Barozzi S, Cittaro D, Kaplunov JM, Bucci G, Dobreva M, Matti V, Beausejour CM, et al. Telomeric DNA damage is irreparable and causes persistent DNA-damage-response activation. Nat Cell Biol 2012; 14:355-65; PMID: 22426077; http://dx.doi.org/10.1038/ncb2466
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 1997; 88:593-602; PMID: 9054499; http://dx.doi.org/10.1016/S0092-8674(00)81902-9
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre' M, Nuciforo PG, Bensimon A, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 2006; 444:638-42; PMID: 17136094; http://dx.doi.org/10.1038/nature05327
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A 1995; 92:9363-7; PMID: 7568133; http://dx.doi.org/10.1073/pnas.92.20.9363
- Hara E, Smith R, Parry D, Tahara H, Stone S, Peters G. Regulation of p16CDKN2 expression and its implications for cell immortalization and senescence. Mol Cell Biol 1996; 16:859-67; PMID: 8622687
- Alcorta DA, Xiong Y, Phelps D, Hannon G, Beach D, Barrett JC. Involvement of the cyclin-dependent kinase inhibitor p16 (INK4a) in replicative senescence of normal human fibroblasts. Proc Natl Acad Sci U S A 1996; 93:13742-7; PMID: 8943005; http://dx.doi.org/10.1073/pnas.93.24.13742
- Narita M, Nunez S, Heard E, Narita M, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 2003; 113:703-16; PMID: 12809602; http://dx.doi.org/10.1016/S0092-8674(03)00401-X
- Dodson H, Bourke E, Jeffers LJ, Vagnarelli P, Sonoda E, Takeda S, Earnshaw WC, Merdes A, Morrison C. Centrosome amplification induced by DNA damage occurs during a prolonged G2 phase and involves ATM. Embo J 2004; 23:3864-73; PMID: 15359281; http://dx.doi.org/10.1038/sj.emboj.7600393
- Ohshima S, Seyama A. Cellular aging and centrosome aberrations. Ann N Y Acad Sci 2010; 1197:108-17; PMID: 20536839; http://dx.doi.org/10.1111/j.1749-6632.2009.05396.x
- Nigg EA, Stearns T. The centrosome cycle: centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol 2011; 13:1154-60; PMID: 21968988; http://dx.doi.org/10.1038/ncb2345
- Gerdes JM, Davis EE, Katsanis N. The vertebrate primary cilium in development, homeostasis, and disease. Cell 2009; 137:32-45; PMID: 19345185; http://dx.doi.org/10.1016/j.cell.2009.03.023
- Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet 2010; 11:331-44; PMID: 20395968; http://dx.doi.org/10.1038/nrg2774
- Kobayashi T, Dynlacht BD. Regulating the transition from centriole to basal body. J Cell Biol 2011; 193:435-44; PMID: 21536747; http://dx.doi.org/10.1083/jcb.201101005
- Hu Q, Milenkovic L, Jin H, Scott MP, Nachury MV, Spiliotis ET, Nelson WJ. A septin diffusion barrier at the base of the primary cilium maintains ciliary membrane protein distribution. Science 2010; 329:436-9; PMID: 20558667; http://dx.doi.org/10.1126/science.1191054
- Ingham PW, Nakano Y, Seger C. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat Rev Genet 2011; 12:393-406; PMID: 21502959; http://dx.doi.org/10.1038/nrg2984
- Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science 2007; 317:372-6; PMID: 17641202; http://dx.doi.org/10.1126/science.1139740
- Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, Reiter JF. Vertebrate smoothened functions at the primary cilium. Nature 2005; 437:1018-21; PMID: 16136078; http://dx.doi.org/10.1038/nature04117
- Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature 2003; 426:83-7; PMID: 14603322; http://dx.doi.org/10.1038/nature02061
- Seeley ES, Nachury MV. The perennial organelle: assembly and disassembly of the primary cilium. J Cell Sci 2010; 123:511-8; PMID: 20144999; http://dx.doi.org/10.1242/jcs.061093
- Conroy PC, Saladino C, Dantas TJ, Lalor P, Dockery P, Morrison CG. C-NAP1 and rootletin restrain DNA damage-induced centriole splitting and facilitate ciliogenesis. Cell Cycle 2012; 11:3769-78; PMID: 23070519; http://dx.doi.org/10.4161/cc.21986
- Mahjoub MR, Stearns T. Supernumerary centrosomes nucleate extra cilia and compromise primary cilium signaling. Curr Biol 2012; 22:1628-34; PMID: 22840514; http://dx.doi.org/10.1016/j.cub.2012.06.057
- Ogura A, Takahashi K. Artificial deciliation causes loss of calcium-dependent responses in Paramecium. Nature 1976; 264:170-2; PMID: 995200; http://dx.doi.org/10.1038/264170a0
- Spektor A, Tsang WY, Khoo D, Dynlacht BD. Cep97 and CP110 suppress a cilia assembly program. Cell 2007; 130:678-90; PMID: 17719545; http://dx.doi.org/10.1016/j.cell.2007.06.027
- Tsang WY, Bossard C, Khanna H, Peranen J, Swaroop A, Malhotra V, Dynlacht BD. CP110 suppresses primary cilia formation through its interaction with CEP290, a protein deficient in human ciliary disease. Dev Cell 2008; 15:187-97; PMID: 18694559; http://dx.doi.org/10.1016/j.devcel.2008.07.004
- Chen Z, Indjeian VB, McManus M, Wang L, Dynlacht BD. CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Dev Cell 2002; 3:339-50; PMID: 12361598; http://dx.doi.org/10.1016/S1534-5807(02)00258-7
- Wong SY, Seol AD, So PL, Ermilov AN, Bichakjian CK, Epstein EH, Jr., Dlugosz AA, Reiter JF. Primary cilia can both mediate and suppress Hedgehog pathway-dependent tumorigenesis. Nat Med 2009; 15:1055-61; PMID: 19701205; http://dx.doi.org/10.1038/nm.2011
- Bishop CL, Bergin AM, Fessart D, Borgdorff V, Hatzimasoura E, Garbe JC, Stampfer MR, Koh J, Beach DH. Primary cilium-dependent and -independent Hedgehog signaling inhibits p16(INK4A). Molecular cell 2010; 40:533-47; PMID: 21095584; http://dx.doi.org/10.1016/j.molcel.2010.10.027
- Leonard JM, Ye H, Wetmore C, Karnitz LM. Sonic Hedgehog signaling impairs ionizing radiation-induced checkpoint activation and induces genomic instability. J Cell Biol 2008; 183:385-91; PMID: 18955550; http://dx.doi.org/10.1083/jcb.200804042
- Chen JK, Taipale J, Cooper MK, Beachy PA. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev 2002; 16:2743-8; PMID: 12414725; http://dx.doi.org/10.1101/gad.1025302
- Loffler H, Lukas J, Bartek J, Kramer A. Structure meets function–centrosomes, genome maintenance and the DNA damage response. Exp Cell Res 2006; 312:2633-40; PMID: 16854412; http://dx.doi.org/10.1016/j.yexcr.2006.06.008
- Dashti M, Peppelenbosch MP, Rezaee F. Hedgehog signalling as an antagonist of ageing and its associated diseases. BioEssays: News Rev Mol, Cell Dev Biol 2012; 34:849-56; PMID: 22903465; http://dx.doi.org/10.1002/bies.201200049
- Zhang H, Herbert BS, Pan KH, Shay JW, Cohen SN. Disparate effects of telomere attrition on gene expression during replicative senescence of human mammary epithelial cells cultured under different conditions. Oncogene 2004; 23:6193-8; PMID: 15195144; http://dx.doi.org/10.1038/sj.onc.1207834
- Kiprilov EN, Awan A, Desprat R, Velho M, Clement CA, Byskov AG, Andersen CY, Satir P, Bouhassira EE, Christensen ST, et al. Human embryonic stem cells in culture possess primary cilia with hedgehog signaling machinery. J Cell Biol 2008; 180:897-904; PMID: 18332216; http://dx.doi.org/10.1083/jcb.200706028
- Goto H, Inoko A, Inagaki M. Cell cycle progression by the repression of primary cilia formation in proliferating cells. Cell Mol Life Sci 2013; 70:3893-905; PMID: 23475109; http://dx.doi.org/10.1007/s00018-013-1302-8
- Seeley ES, Carriere C, Goetze T, Longnecker DS, Korc M. Pancreatic cancer and precursor pancreatic intraepithelial neoplasia lesions are devoid of primary cilia. Cancer research 2009; 69:422-30; PMID: 19147554; http://dx.doi.org/10.1158/0008-5472.CAN-08-1290
- Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell 2007; 129:1351-63; PMID: 17604723; http://dx.doi.org/10.1016/j.cell.2007.04.035
- Inoko A, Matsuyama M, Goto H, Ohmuro-Matsuyama Y, Hayashi Y, Enomoto M, Ibi M, Urano T, Yonemura S, Kiyono T, et al. Trichoplein and Aurora A block aberrant primary cilia assembly in proliferating cells. J Cell Biol 2012; 197:391-405; PMID: 22529102; http://dx.doi.org/10.1083/jcb.201106101
