Abstract
The tripartite motif protein family (TRIM) constitutes a class of immune-regulated proteins with antiviral, immune, cancer, and other properties reminiscent of those ascribed to autophagy. We show that TRIMs have dual roles in autophagy: as regulators and as cargo receptors. As regulators, TRIMs nucleate the core autophagy machinery by acting as platforms that assemble ULK1 and BECN1 into a functional complex in preparation for autophagy. TRIMs also act as novel selective autophagy receptors as exemplified by TRIM5/TRIM5α, a known HIV-1 restriction factor with a hitherto poorly defined mode of action. TRIM5 recognizes and targets HIV-1 for autophagic destruction. TRIM5 interactions with mammalian Atg8 proteins are required for this effector function. This establishes TRIM family members as regulators of autophagy, explains the antiretroviral mechanism of TRIM5, and defines a new basis for selective autophagy.
Autophagy is intimately integrated with immune systems in metazoans through engagement of nearly all major classes of pattern recognition receptors (PRRs). Among PRRs, one notable exception is the tripartite motif class of proteins, consisting of more than 70 members in humans. Despite some indications of individual TRIMs being connected to autophagy, the TRIM family surprisingly has not been systematically analyzed for a role in this process. We tested the hypothesis that, akin to other classes of PRRs, the TRIM family may play a general role in autophagy. We performed an siRNA screen examining the effects of TRIM knockdowns on autophagy using GFP-LC3 puncta as a readout. Remarkably, roughly half of the TRIMs tested affected induced autophagy, and additional TRIMs were found to regulate basal autophagy. These data indicated that autophagy regulation is a conserved feature among TRIMs, and invited in-depth studies to determine their mode of action.
For detailed analysis of how a TRIM participates in autophagy, we chose to focus first on TRIM5, a well-known retroviral restriction factor. TRIM5 knockdown inhibited autophagy induction as measured by LC3-II levels and by LC3 puncta abundance. Conversely, overexpression of TRIM5 induced autophagy. As our data indicated that TRIM5 is involved in autophagy initiation, we tested whether this and other TRIMs interacted with either ULK1 or BECN1, 2 flagship members of the key autophagy regulatory complexes. Of the 6 TRIMs tested (5, 6, 17, 22, 49, and 55), all but TRIM55 were in protein complexes with both ULK1 and BECN1. As BECN1 activity is directly regulated via an activating phosphorylation by ULK1, we considered a model in which TRIMs act as platforms for the gathering or generation of active ULK1 and BECN1. Accordingly, TRIM5 was preferentially in a complex with active phospho-ULK1 (p-Ser-317). Furthermore, the expression of TRIMs 5, 6, 17, 22, and 49 all promoted the formation of multimolecular complexes containing both ULK1 and BECN1. Finally, TRIM expression promoted BECN1 activation, as it resulted in dissociation from BECN1 of its negative regulators BCL2 and TAB2. Thus, TRIMs regulate autophagy by serving as platforms for the nucleation and activation of the autophagy initiation machinery.
In addition to regulating autophagy, we found that TRIMs can also act as selective autophagy receptors. We noticed the presence of LC3-interacting region (LIR)-like motifs in many TRIMs including TRIM5. We therefore tested if TRIMs could interact with mammalian paralogs of yeast Atg8 (mAtg8s). We found that all 7 TRIMs tested directly interacted with at least one mAtg8 (GABARAP). Interestingly, included in this list are 2 TRIMs that are ‘negative’ in our screen, hinting that our screen likely underestimated the extent to which TRIMs are connected to autophagy. Further tests with TRIM5 showed that it also colocalized with autophagosomes and cofractionated with lipidated LC3. Based on these results, we tested whether TRIM5 could direct the autophagic degradation of incoming retroviral cores. The rhesus TRIM5 (RhTRIM5) SPRY domain recognizes human immunodeficiency virus 1 (HIV-1) capsid composed of the viral protein p24. HIV-1 capsid protein p24 is a substrate for lysosomal degradation, because inhibition of lysosomal proteases protected it in rhesus FRhK4 cells early during infection. Capsid p24 degradation increased upon induction of autophagy by starvation and was abrogated, under both basal and starvation conditions, by knockdowns of ATG7, BECN1, SQSTM1/p62, and TRIM5 in FRhK4 and in primary rhesus CD4+ T cells. We next sought to determine if this was dependent on the specific interaction between TRIM5 and p24 as its protein target. For this, we employed a feature of rhesus TRIM5, which recognizes HIV-1 capsid but not the SIV retroviral capsid. Autophagy, induced by starvation, resulted in a reduced output of virally-encoded luciferase only with HIV, but not with SIV in an ATG7-, BECN1-, SQSTM1- and TRIM-dependent manner. Thus, mobilization of the viral target for degradation by the autophagic apparatus correlates with the known binding specificity of TRIM5.
Further studies have demonstrated that TRIM5 is a bona fide autophagic receptor, as its ability to deliver HIV-1 capsid for autophagic degradation is dependent on its interaction with mAtg8s. Using a peptide array, we identified 2 adjacent LIR motifs in TRIM5. When these motifs were mutated, TRIM5 lot binding to mAtg8s and, whereas expression of rhesus TRIM5 in human cells induced HIV-1 p24 capsid degradation, the expression of the LIR mutant of RhTRIM5 did not restrict the virus.
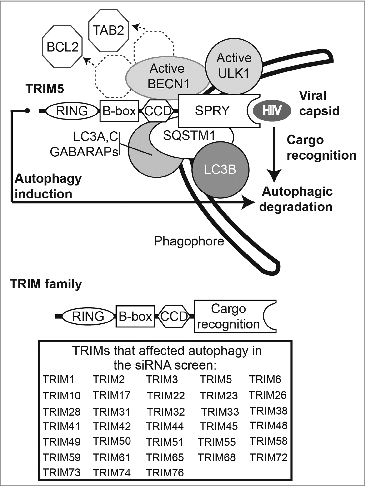
In conclusion, our findings demonstrate that TRIM proteins as a family broadly affect autophagy. The majority of TRIMs studied act as organizing platforms to orchestrate ULK1 and BECN1 action. In its receptor role, TRIM5 directly recognizes, without a need for ubiquitin-tagging, its cognate retrovirus capsid and orchestrates its autophagic degradation, thus protecting cells against HIV-1 infection. Based on these features, TRIMs embody in a single protein entity several aspects of selective autophagy: they recognize the target, organize autophagy initiation machinery, and induce autophagy. We propose a model () in which TRIM5 and several other TRIMs assemble mammalian-like phagophore assembly sites directing a subset of selective autophagy.
Figure 1. Top, TRIM5 promotes autophagy by acting as a platform for the assembly of active ULK1 and BECN1 complexes and rhesus (but not human) TRIM5 delivers incoming HIV-1 capsids for autophagic degradation by directly binding both the viral capsid and mammalian Atg8s. Assembly of ULK1, BECN1, and mammalian Atg8s is a conserved feature in most TRIMs. We propose that TRIMs use their SPRY or other cargo recognition domains for the autophagic targeting of a wide variety of microbial and endogenous cellular substrates. Shown is a generic TRIM domain organization; the table lists TRIMs that affect autophagy in our siRNA screen.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
