Abstract
One of the limitations of the use of phage antibody libraries in high throughput selections is the production of sufficient phage antibody library at the appropriate quality. Here, we successfully adapt a bioreactor-based protocol for the production of phage peptide libraries to the production of phage antibody libraries. The titers obtained in the stirred-tank bioreactor are 4 to 5 times higher than in a standard shake flask procedure, and the quality of the phage antibody library produced is indistinguishable to that produced using standard procedures as assessed by Western blotting and functional selections. Availability of this protocol will facilitate the use of phage antibody libraries in high-throughput scale selections.
Abbreviations
| scFv | = | single chain Fv |
| VH | = | Variable Heavy |
| VL | = | Variable Light |
| g3p | = | gene 3 protein |
| tTG2 | = | tissue transglutaminase 2 |
Introduction
Phage displayCitation1 is one of the most successful platforms for the isolation of peptide or antibody ligands for potentially any target molecule, with phage antibody librariesCitation2,Citation3 having the potential to provide hundreds of unique antibodies per target,Citation4 especially when combined with yeast display.Citation5 Once selected, the affinities of antibodies or peptides selected by phage display can be matured relatively easily.Citation6-Citation9 Most antibody libraries use single-chain variable fragments (scFvs)Citation10 as the antibody format, in which a flexible linker joins the VH and VL chains; or Fabs, where VH-CH1 and VL-CL associate non-covalently in the periplasmic space. Antibody libraries can be created either syntheticallyCitation11-Citation16 by introducing oligonucleotide encoded diversity into frameworks with desirable properties, or by harvesting natural diversity from human lymphocytes by PCR.Citation2,Citation17-Citation19
Most phage antibody libraries have been created by cloning large numbers of different antibody genes upstream of gene 3 and using phageCitation20 or phagemidCitation3,Citation14,Citation17,Citation21-Citation24 vectors as the display vehicles. In general, phagemid vectors are preferred because cloning is more straightforward and they are more stable. However, whereas display using phage is multivalent (up to 5 copies), that with phagemids is monovalent, with only 1% of phagemid particles actually displaying antibodies.Citation25 Although new approaches for generation of phage(mid) antibody libraries, such as monodisperse emulsion,Citation26 have been described, production is usually carried out in shake flasks, with the obtained phage titer from one 2.5 L flask sufficient for approximately 50 selections. Recently, a number of high throughput antibody selection platforms based on phage display have been described,Citation27-Citation31 where selections are carried out against numerous different antigens in parallel. These approaches require a large amount of high quality phage antibody library as starting material, the generation of which presents some challenges, including appropriate expression of functional antibody, purification of phage particles, quality control of displayed antibody in terms of actual diversity, effective display of the scFv and the ability to select antibodies against targets.
Fermentation technology has been used to produce large amounts of functional phage-displayed peptide libraries.Citation32 Inspired by this study, we tested the methodology for the production of our antibody phage library.Citation3,Citation33 We demonstrate that this method can be used to produce large amounts of functional phage antibody particles from a single preparation, resolving one of the bottlenecks to high throughput phage antibody selections.
Results
Media comparison for phage library production
Before attempting phage antibody production in the bioreactor, 2 different media were investigated for the expression of our antibody phage library. This was based on our standard protocols, or those previously used to produce peptide phage libraries.Citation32 After reaching mid-log phase, bacteria were infected with our recombined antibody phage library, followed by helper phage infection, which supplies all the other proteins required for assembly of phage particles displaying and encoding scFvs. The media were first tested in flasks. Bacterial growth (at 37°C before and 30°C after infection) and phage multiplicity of infection were kept constant for the media tested and a buffering solution used to avoid pH fluctuation. The bacteriophage-infected cells grew exponentially until the plateau phase, reached after approximately 12 hours growth when 2xYT was used, and around 10 h when NZY was used ().
Figure 1. Two different media were tested in flasks for the production of antibody phage library. After double infection, bacteria reached the plateau phase more rapidly when NZY medium was used (A), which was associated with a higher phage titer (B). All the experiments were conducted in triplicate and +/− standard deviations reported.
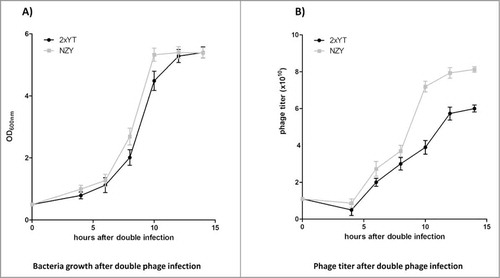
The phage titer started to increase when bacteria restarted exponential growth, at around 6 h after infection, and approached plateau at 10 hours for NZY and 12 hours for 2xTY, with peak levels at approximately 14 hours for both media (). Final bacteriophage titers at the end of growth were 6 × 1010 phage particle/ml for 2xYT and 8 × 1010 phage/ml for NZY.
Phage production using a stirred-tank bioreactor
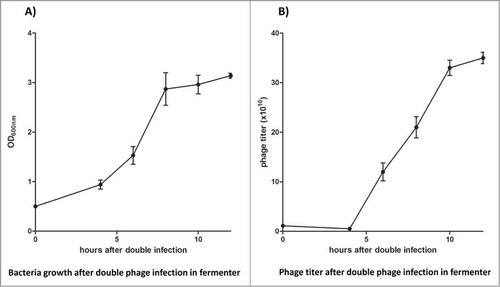
At the end of growth in flasks, both media reached similar phage titers, but NZY medium had the advantage that phage production appeared to be faster, with plateau production starting earlier. For this reason we decided to use 5 L of NZY in our bioreactor experiment. The same time points were used as those in the preliminary data obtained in the flask condition - to control both bacterial growth and phage production. Using the stirred-tank bioreactor, we noticed slower bacterial growth, with a final density corresponding to an OD600 of 3 (), which was lower than the one obtained using flasks. However, the amount of phage produced by the end of the experiment was 3.5 × 1011 phage/ml (), 4–5 times higher than the titer obtained in flasks.
Figure 2. NZY medium was used in the production of phage antibody library in the stirred-tank bioreactor. Bacterial growth (A) and phage titer (B) were monitored, with a final phage particle production almost 5 times larger than that obtained with flask growth. All the experiments were conducted in triplicate and +/− standard deviations reported.
Phage quality control
Two hundred individual clones from each of the 3 different library preparations (2 with standard methods but different media, and one using the bioreactor) were analyzed for the presence of full-length scFvs by PCR (). We found that 15% (2xYT library) and 18% (NZY library) of the clones from the 2 libraries generated by standard protocols had deleted scFv genes, with a similar result (19%) obtained from the stirred-tank bioreactor grown library. These deletions are likely to have occurred during the library construction, probably as a result of spurious priming during PCR assemblyCitation3, and not due to the growing condition of the amplified library, and reflect the levels of deletion we usually see. To rapidly check the diversity of these amplified clones, the PCR-amplified scFvs were characterized by fingerprinting with the restriction enzyme, BstNI. The results obtained with the different growth-conditions were essentially identical, with different fingerprint patterns representing different VH and VL gene usage in the different library preparations ().
Figure 3. Phage antibody library quality for the 3 libraries was determined by: (A) PCR reaction to estimate the ratio of full length scFv (∼800bp) to deleted fragments (∼400bp); (B) fingerprinting to assess the diversity of the different single clones; (C) Western blotting to evaluate scFv display levels on the surface of phage. For panel (a) and (b) PCR and fingerprinting form clones of the stirred-tank bioreactor-library are shown as an example.
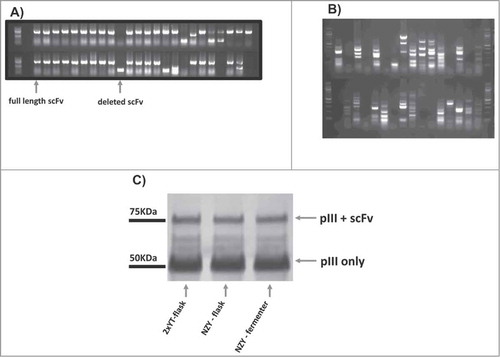
The scFv display level was examined by western blot () using the SV5 anti-tag antibodyCitation34, which recognizes the SV5 tag placed between the displayed scFv and p3. The assay gives an assessment of the portion of recombinant p3, which is full length and displays scFv (p3+scFv) compared to the amount of recombinant p3 derived from the proteolytic degradation of g3p+scFv. The results show that the display levels obtained from the different preparation are essentially identical, as shown by the intensity of the p3+scFv band and p3 band.
Phage antibody selection performance comparison
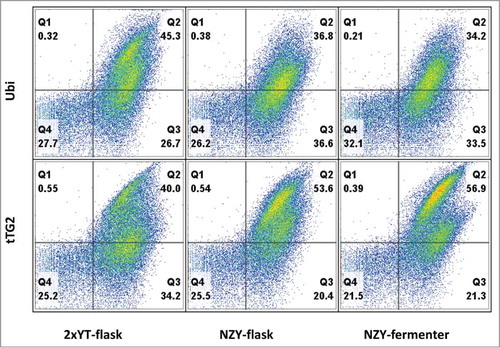
The final quality-control test consisted in challenging the 3 antibody libraries in our standard antibody selection procedure. The 3 libraries were selected on 2 different antigens, ubiquitin and tTG2. We applied a recently developed antibody selection pipeline, based on the combination of phage and yeast displayCitation5 to qualitatively estimate the ability of the produced phage antibody libraries to select functional antibodies against specific targets. After 2 rounds of phage selection, the total outputs were subcloned into a yeast display vector and sorted by flow cytometry twice more. The populations obtained were analyzed and compared by flow cytometry, which allows the analysis of many different antibodies in a single experiment.Citation5 Yeast clones displaying scFv that are displayed and bind to their target can be visualized in the top right quadrant. The percentage of binders is similar in the 3 different libraries (34–45% for ubiquitin, and 40–57% for tTG2) (), with little significant difference between the shape of the clouds in the different selections, indicating that antibodies with similar properties, in terms of display levels and binding abilities, were selected.
Figure 4. The different libraries were challenged with 2 antigens (ubiquitin – Ubi, tissue transglutaminase 2 – tTG2), by performing 2 rounds of phage selection followed by subcloning the phage selected outputs into a yeast display vector and sorting by flow cytometry 2 additional times. After analyzing the yeast populations, with the yeast clones displaying scFv binding to their target in the top right quadrant, it is possible to appreciate the similar percentage of binders in the 3 different libraries (34–45% for ubiquitin, and 40–57% for tTG2).
Discussion
Antibody phage libraries are widely used to select antibodies against diverse targets. This method can be very effectively scaled up and used for high-throughput approaches,Citation27-Citation31 with the antibodies obtained comparable to those obtained by immunization.Citation27,Citation28 The ability to carry out industrial-scale selections against different targets in parallel depends, however, on the availability of suitable amounts of phage antibody library. Here, we show that effective phage antibody libraries can be produced in large volumes using bioreactors, and their quality, as assessed by titer, display level and selection success, is indistinguishable from libraries made in shake flasks. The amount of phage antibody library produced in a single 5 l stirred-tank bioreactor run is equivalent to the production of approximately 20 2.5 l shake flasks, - sufficient for 1,000 selections, - but the bioreactor-based protocol requires substantially less effort. The use of bioreactors also facilitates standardization of the protocol, ensuring minimal lot-to-lot variation in the quality of antibody library produced.
Although the use of fermenters to produce large quantities of phage antibody libraries has not been previously described, phage peptide libraries have been produced using this approach.Citation32 Apart from the difference in size of the displayed molecule (short peptide versus 25 kDa scFv), the biggest difference between display of peptides or antibodies is the nature of the vector used. Peptide display usually uses filamentous phage vectors, in which it is sufficient to grow bacteria containing the vector for phage displaying peptides to be produced. In the case of antibody libraries, phagemid vectors are usually used, and the vector alone is unable to generate phage particles unless the bacteria carrying them has been co-infected with helper phage, which provides all the other proteins needed for phage particle assembly. Here we show that, notwithstanding these differences, the fermentation conditions described for peptide phage library production are applicable to phagemid antibody libraries as well, with titers 4-5 fold higher than those obtained by growth in flasks. Although further experimentation may identify conditions that provide even higher titers, those we describe here should be suitable for most purposes in which large amounts of high quality phage antibody library is required.
Material and Methods
Antigen
Ubiquitin was purchased from Sigma and tissue transglutaminase 2 (tTG2) from Zedira. Both were biotinylated using the Pierce Biotinylation Kit (Pierce) to a final concentration of 1 mg/ml. The effective biotinylation of more than 95% of the proteins was checked by western blot and ELISA (data not shown).
Phage library, bacterial strain and media
The phage antibody library used in this study was previously described.Citation3 Briefly, the phage antibody library requires infection with helper phage and propagation in F-pilus bacteria to allow the coupling of a unique scFv with a single phage particle.
E.coli DH5αF’ [F’/endA1 hsdR17(rK- mK+) supE44 thi-1 recA1 gyrA (Nalr) relA1 Δ (lacZYA-argF)U169 deoR (ϕ80dlacΔ(lacZ)M15)] was used to propagate and express bacteriophage.
Two different media, 2xYT and NZY were compared for phage production. 2xYT consists of 31 g/l of Difco™ 2xYT Broth Formulation (BD Bioscience) in 1 l final volume (900 ml of distilled water and 100 ml of 1 M HEPES solution (Fisher)) buffered at pH 7.4 (with 1 M of NaOH); NZY was prepared following Grieco et alCitation32: 10 g of NZ-Amine (Sigma-Aldrich), 5 g yeast extract (BD Bioscience), 5 g NaCl in 1 l final volume (900 ml distilled water and 100 ml of 1M HEPES solution) and pH adjusted to 7.4 (with 1M of NaOH).
2xYT and NZY plates were prepared by adding 10 g/l of select agar (Fisher Scientific), 1% final concentration of glucose and the required antibiotic.
Small-scale phage production
A single colony of DH5αF’ E.coli was picked from a 2xYT agar plate and added to 10 ml of either 2xYT or NZY broth and grown overnight (ON) at 30°C. The ON inoculum was added to 1000 ml of 2xYT (or NZY) divided in 2 (500 ml each) 2.5 l flasks (Thomson Instruments) containing 3 % glucose and grown to OD600 0.3 at 37°C shaking at 250 rpm. Phage libraryCitation3 was added at multiplicity of infection (MOI - phages : bacteria) of 1:1.25 diluted in 20 ml (pre-warmed) of 2xYT (or NZY) at 37°C for 30 min to allow phage infection. Afterward, helper phage M13K07 was added at a MOI of 10:1 in a total volume of 20 ml pre warmed 2xYT (or NZY) at 37°C for 45 min to finalize the double infection. Bacteria were pelleted by centrifugation and re-suspended in 1,000 ml of carb/kan (50 mg/ml carbenicillin, 25 mg/ml kanamycin) 2xYT (or NZY) to allow the growth of only double-infected bacteria. Cells were grown at 30°C for 14 h. To perform phage selection, phage were purified by PEG-precipitation: the culture was centrifuged at 10,000 rpm and the supernatant PEG precipitated twice by adding 20 % volume of 20 % PEG 8,000, 2.5M NaCl. Phage were resuspended in a final volume of 20 ml PBS supplemented with glycerol for storage and protease inhibitor cocktail to prevent the cleavage of the displayed scFvs.
Bioreactor-based cultivation
The cultivation was performed in 4.5 l of NZY medium supplemented with 500 ml of 1 M HEPES - final 5 l solution buffered at pH 7.4 - using a BioFlo 2000 Fermenter (New Brunswick Scientific Co). The medium, poured into a 10 l vessel, was autoclaved and cooled to a working temperature of 37°C. In order to start cultivation, the bioreactor was set up with constant temperature at 37°C, agitation at 150 rpm, and an air-supply with a flow-rate of 5 l/min.
Fifty ml of E. coli DH5αF’ cells prepared by ON culture were added. The cells were grown to mid-log phase (OD600 of 0.3), the agitation was lowered to 50 rpm and the phage library - at the same MOI used for flask production (1:1.25) - , diluted to 25 ml, was added to the cell culture. The cultivation was continued for 30 min. Then, after adding 60 ml of helper phage (5 × 1012 phage/ml, final MOI 10:1), infected bacteria were grown for another 45 min and the 2 antibiotics were added: carbenicillin to a final concentration 50 μg/ml for selection of the phage library and kanamycin to a final 25 μg/ml concentration to select for the helper phage. The temperature was then adjusted to 30°C and the culture was agitated at 150 rpm. Cultivation was continued for an additional 14 hours cell growth and titration of bacteriophage were monitored and performed as described below.
Measurement of cell growth and titration of bacteriophage
Cell growth was checked by collecting samples after double infection at 0, 4, 6, 8, 10, 12 and 14 hours after infection and by measuring OD600 with a spectrometer (BD Bioscience). At the same time-points samples were collected and centrifuged at 5,000 rpm at room temperature (RT), to remove bacteria, and 10 μl of phage-containing culture supernatant were added to 990 μl of medium in a microtiter well. A 10-fold dilution series was carried out in the microtiter plate in order to determine both carbenicillin (phagemid) and kanamycin (helper phage) titers. 100 μl of DH5αF’ grown to 0.5 OD600 at 37°C was added to each well and phage infection was allowed to occur for 30 min at 37°C. Five μl of the infected cells from each well were spotted on agar plates containing carbenicillin and glucose and incubated at 30°C ON. The phage titer was calculated from the dilution with the highest number of countable colonies.
Phage library quality controls
Insert size was checked by colony PCR by using the primers pDpH-5’ and pDpH-3’Citation3. Three μl of PCR reaction were run on an agarose gel to assess the size of the amplicon. The rest of the reaction was digested with BstNI (NEB) restriction enzyme to perform fingerprinting analysis to determine the diversity of the amplified scFvs.
Western blot analysis of phage display: after PEG precipitation, 5 × 1011 phage particles from every single preparation (flask and stirred-tank bioreactor) were electrophoresed using the NuPAGE gel system (Life Technologies). After electrophoresis, the proteins were transferred to nitrocellulose membrane using a semidry blot apparatus. The membrane was blocked with 3 % milk PBST for 1 h at RT. This was then incubated in 1 mg/ml of anti-SV5-AP conjugated antibody in 2 % milk PBST for 1 h at RT. The blot was washed 2xPBST and 1xPBS, 5 min each. After washing, the AP activity was detected using NBT-BCIP substrate (Sigma).
Antibody Selection Evaluation
The produced phage antibody libraries were used to select anti-ubiquitin and anti-tTG2 antibodies with an automated Kingfisher magnetic bead system (Thermo Lab Systems), allowing selection to be carried out in solution as previously described.Citation35 An excess of biotinylated antigen was used to saturate 2 × 107 streptavidin magnetic beads (Dynabeads M-280) which were used to capture specific phage. The second round section outputs were subcloned into a yeast display vector as described by Ferrara et al.Citation5. The yeast mini-libraries were further enriched by one or 2 rounds of sorting by flow cytometry using published methods.Citation36 Briefly, 2 × 10Citation6 yeast cells were washed and resuspended in 100 μl of wash buffer containing 100 nM of biotinylated antigens (ubiquitin and tTG2). Cells were labeled with streptavidin-AlexaFluor-633 to detect binding of biotinylated targets and 1 μg of anti-SV5-PE to assess scFv display levels. Flow cytometry was performed using the FACSAria (Becton Dickinson), with yeast showing antigen and SV5 binding sorted. Collected cells were grown overnight at 30°C and induced for the next round of sorting. BDiva software was used for flow cytometric analysis.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors’ Contributions
FF and AMB designed the experiments. FF and CK performed the cell growth experiments, LAN conducted the library characterization experiments. FF and AMB wrote the manuscript.
Funding
This work was supported by Los Alamos National Laboratory and New Mexico Consortium through a NIH U54 Grant “Technology Development for New Affinity Reagents Against the Human Proteome (U54) RFA-RM-10–018," grant number 1-U54-DK093500-01.
References
- Scott JK, Smith GP. Searching for peptide ligands with an epitope library. Science 1990; 249:386; PMID:1696028; http://dx.doi.org/10.1126/science.1696028
- Marks JD, Hoogenboom HR, Bonnert TP, McCafferty J, Griffiths AD, Winter G. By-passing immunization. Human antibodies from V-gene libraries displayed on phage. J Mol Biol 1991; 222:581; PMID:1748994; http://dx.doi.org/10.1016/0022-2836(91)90498-U
- Sblattero D, Bradbury A. Exploiting recombination in single bacteria to make large phage antibody libraries. Nat Biotechnol 2000; 18:75; PMID:10625396; http://dx.doi.org/10.1038/71958
- Edwards BM, Barash SC, Main SH, Choi GH, Minter R, Ullrich S, Williams E, Du Fou L, Wilton J, Albert VR, et al. The remarkable flexibility of the human antibody repertoire; isolation of over one thousand different antibodies to a single protein, BLyS. J Mol Biol 2003; 334:103; PMID:14596803; http://dx.doi.org/10.1016/j.jmb.2003.09.054
- Ferrara F, Naranjo LA, Kumar S, Gaiotto T, Mukundan H, Swanson B, Bradbury AR. Using phage and yeast display to select hundreds of monoclonal antibodies: application to antigen 85, a tuberculosis biomarker. PLoS One 2012; 7:e49535; PMID:23166701; http://dx.doi.org/10.1371/journal.pone.0049535
- Schier R, McCall A, Adams GP, Marshall KW, Merritt H, Yim M, Crawford RS, Weiner LM, Marks C, Marks JD. Isolation of picomolar affinity anti-c-erbB-2 single-chain Fv by molecular evolution of the complementarity determining regions in the center of the antibody binding site. J Mol Biol 1996; 263:551; PMID:8918938; http://dx.doi.org/10.1006/jmbi.1996.0598
- Low NM, Holliger PH, Winter G. Mimicking somatic hypermutation: affinity maturation of antibodies displayed on bacteriophage using a bacterial mutator strain. J Mol Biol 1996; 260:359; PMID:8757799; http://dx.doi.org/10.1006/jmbi.1996.0406
- Marks JD, Griffiths AD, Malmqvist M, Clackson TP, Bye JM, Winter G. By-passing immunization: building high affinity human antibodies by chain shuffling. Biotechnology (N Y) 1992; 10:779; PMID:1368267; http://dx.doi.org/10.1038/nbt0792-779
- Gram H, Marconi LA, Barbas CF, 3rd, Collet TA, Lerner RA, Kang AS. In vitro selection and affinity maturation of antibodies from a naive combinatorial immunoglobulin library. Proc Natl Acad Sci U S A 1992; 89:3576; PMID:1565653; http://dx.doi.org/10.1073/pnas.89.8.3576
- Huston JS, Levinson D, Mudgett-Hunter M, Tai MS, Novotny J, Margolies MN, Ridge RJ, Bruccoleri RE, Haber E, Crea R, et al. Protein engineering of antibody binding sites: recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc Natl Acad Sci U S A 1988; 85:5879; PMID:3045807; http://dx.doi.org/10.1073/pnas.85.16.5879
- Hoogenboom HR, Winter G. By-passing immunisation. Human antibodies from synthetic repertoires of germline VH gene segments rearranged in vitro. J Mol Biol 1992; 227:381; PMID:1404359; http://dx.doi.org/10.1016/0022-2836(92)90894-P
- Krebs B, Rauchenberger R, Reiffert S, Rothe C, Tesar M, Thomassen E, Cao M, Dreier T, Fischer D, Hoss A, et al. High-throughput generation and engineering of recombinant human antibodies. J Immunol Methods 2001; 254:67; PMID:11406154; http://dx.doi.org/10.1016/S0022-1759(01)00398-2
- Birtalan S, Fisher RD, Sidhu SS. The functional capacity of the natural amino acids for molecular recognition. Mol Bio Systems 2010; 6:1186; PMID:20383388; http://dx.doi.org/10.1039/b927393j
- Birtalan S, Zhang Y, Fellouse FA, Shao L, Schaefer G, Sidhu SS. The intrinsic contributions of tyrosine, serine, glycine and arginine to the affinity and specificity of antibodies. J Mol Biol 2008; 377:1518; PMID:18336836; http://dx.doi.org/10.1016/j.jmb.2008.01.093
- Fellouse FA, Esaki K, Birtalan S, Raptis D, Cancasci V J, Koide A, Jhurani P, Vasser M, Wiesmann C, Kossiakoff AA, et al. High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries. J Mol Biol 2007; 373:924; PMID:17825836; http://dx.doi.org/10.1016/j.jmb.2007.08.005
- Sidhu SS, Kossiakoff AA. Exploring and designing protein function with restricted diversity. Curr Opin Chem Biol 2007; 11:347; PMID:17500026; http://dx.doi.org/10.1016/j.cbpa.2007.05.001
- Vaughan TJ, Williams AJ, Pritchard K, Osbourn JK, Pope AR, Earnshaw JC, McCafferty J, Hodits RA, Wilton J, Johnson KS. Human antibodies with sub-nanomolar affinities isolated from a large non-immunized phage display library. Nat Biotechnol 1996; 14:309; PMID:9630891; http://dx.doi.org/10.1038/nbt0396-309
- Lloyd C, Lowe D, Edwards B, Welsh F, Dilks T, Hardman C, Vaughan T. Modelling the human immune response: performance of a 1011 human antibody repertoire against a broad panel of therapeutically relevant antigens. Protein Eng Des Sel 2009; 22:159; PMID:18974080; http://dx.doi.org/10.1093/protein/gzn058
- Schofield DJ, Pope AR, Clementel V, Buckell J, Chapple S, Clarke KF, Conquer JS, Crofts AM, Crowther SR, Dyson MR, et al. Application of phage display to high throughput antibody generation and characterization. Genome Biol 2007; 8:R254; PMID:18047641; http://dx.doi.org/10.1186/gb-2007-8-11-r254
- Huie MA, Cheung MC, Muench MO, Becerril B, Kan YW, Marks JD. Antibodies to human fetal erythroid cells from a nonimmune phage antibody library. Proc Natl Acad Sci USA 2001; 98:2682; PMID:11226299; http://dx.doi.org/10.1073/pnas.051631798
- de Haard HJ, van Neer N, Reurs A, Hufton SE, Roovers RC., Henderikx P, de Bruine AP, Arends JW, Hoogenboom HR. A large non-immunized human Fab fragment phage library that permits rapid isolation and kinetic analysis of high affinity antibodies. J Biol Chem 1999; 274:18218; PMID:10373423; http://dx.doi.org/10.1074/jbc.274.26.18218
- Sheets MD, Amersdorfer P, Finnern R, Sargent P, Lindqvist E, Schier R, Hemmingsen G, Wong C, Gerhart JC, Marks JD. Efficient construction of a large nonimmune phage antibody library: the production of high-affinity human single-chain antibodies to protein antigens. Proc Natl Acad Sci USA 1998; 95:6157; PMID:9600934; http://dx.doi.org/10.1073/pnas.95.11.6157
- Knappik A, Ge L, Honegger A, Pack P, Fischer M, Wellnhofer G, Hoess A, Wolle J, Pluckthun A, Virnekas B. Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and CDRs randomized with trinucleotides. J Mol Biol 2000; 296:57; PMID:10656818; http://dx.doi.org/10.1006/jmbi.1999.3444
- Gilbreth RN, Esaki K, Koide A, Sidhu SS, Koide S. A dominant conformational role for amino acid diversity in minimalist protein-protein interfaces. J Mol Biol 2008; 381:407; PMID:18602117; http://dx.doi.org/10.1016/j.jmb.2008.06.014
- Clackson T, Wells JA. In vitro selection from protein and peptide libraries. Trends Biotechnol 1994; 12:173; PMID:7764900; http://dx.doi.org/10.1016/0167-7799(94)90079-5
- Matochko WL, Ng S, Jafari MR, Romaniuk J, Tang SK, Derda R. Uniform amplification of phage display libraries in monodisperse emulsions. Methods 2012; 58:18; PMID:22819853; http://dx.doi.org/10.1016/j.ymeth.2012.07.012
- Colwill K, Persson H, Jarvik NE, Wyrzucki A, Wojcik J, Koide A, Kossiakoff AA, Koide S, Sidhu S, Dyson MR, et al. A roadmap to generate renewable protein binders to the human proteome Nat Methods 2011; 8:551; PMID:21572409; http://dx.doi.org/10.1038/nmeth.1607
- Pershad K, Pavlovic JD, Graslund S, Nilsson P, Colwill K, Karatt-Vellatt A, Schofield DJ, Dyson MR, Pawson T, Kay BK, McCafferty J. Generating a panel of highly specific antibodies to 20 human SH2 domains by phage display. Protein Eng Des Sel 2010; 23:279; PMID:20164216; http://dx.doi.org/10.1093/protein/gzq003
- Mersmann M, Meier D, Mersmann J, Helmsing S, Nilsson P, Graslund S, Colwill K, Hust M, Dubel S. Towards proteome scale antibody selections using phage display. New Biotechnol 2010; 27:118; PMID:19883803; http://dx.doi.org/10.1016/j.nbt.2009.10.007
- Hallborn J, Carlsson R. Automated screening procedure for high-throughput generation of antibody fragments. Biotechniques 2002; 33:Suppl., 30-37; PMID:12514927
- Lou J, Marzari R, Verzillo V, Ferrero F, Pak D, Sheng M, Yang C, Sblattero D, Bradbury A. Antibodies in haystacks: how selection strategy influences the outcome of selection from molecular diversity libraries, J Immunol Methods 2001; 253:233; PMID:11384684; http://dx.doi.org/10.1016/S0022-1759(01)00385-4
- Grieco SH, Lee S, Dunbar WS, MacGillivray, RT, Curtis SB. Maximizing filamentous phage yield during computer-controlled fermentation. Bioprocess Biosystems Eng 2009; 32:773; PMID:19221805; http://dx.doi.org/10.1007/s00449-009-0303-3
- Sblattero D, Lou J, Marzari R, Bradbury A. In vivo recombination as a tool to generate molecular diversity in phage antibody libraries. Rev Mol Biotech 2001; 74:303; PMID:11526909; http://dx.doi.org/10.1016/S1389-0352(01)00022-8
- Randall RE, Young DF. Comparison between parainfluenza virus type 2 and simian virus 5: monoclonal antibodies reveal major antigenic differences. J Gen Virol 1988; 69 ( Pt 8):2051; PMID:2841416; http://dx.doi.org/10.1099/0022-1317-69-8-2051
- Marks JD, Bradbury A. Selection of human antibodies from phage display libraries. Methods Mol Biol 2004; 248:161; PMID:14970495
- Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol 1997; 15:553; PMID:9181578; http://dx.doi.org/10.1038/nbt0697-553
