Abstract
Extra-intestinal pathogenic Escherichia coli (ExPEC) are a significant cause of urinary tract infections and bacteraemia worldwide. Currently no single virulence factor or ExPEC lineage has been identified as the sole contributor to severe extra-intestinal infection and/or urosepsis. Galleria mellonella has recently been established as a simple model for studying the comparative virulence of ExPEC. In this study we investigated the virulence of 40 well-characterized ExPEC strains, in G. mellonella, by measuring mortality (larvae survival), immune recognition/response (melanin production) and cell damage (lactate dehydrogenase production). Although mortality was similar between urinary and bloodstream isolates, it was heightened for community-associated infections, complicated UTIs and urinary-source bacteraemia. Isolates of ST131 and those possessing afa/dra, ompT and serogroup O6 were also associated with heightened virulence.
Abbreviations
| Abx | = | antibiotics |
| CFU | = | colony forming unit |
| ExPEC | = | extra-intestinal pathogenic E. coli |
| LDH | = | lactate dehydrogenase |
| MLST | = | multi-locus sequence type |
| OD | = | optical density |
| ST | = | sequence type |
| UTI | = | urinary tract infection |
| VF | = | virulence factor |
Introduction
Extra-intestinal pathogenic Escherichia coli (ExPEC) are well known as a significant cause of urinary tract infection (UTI) and bloodstream infections worldwide.Citation1-3 Since their designation by Russo and Johnson 14 years ago, various characteristics of the ExPEC population have been described.Citation4 Although 7 distinct phylogenetic groups (phylogroups) have been defined (A, B1, B2, C, D, E and F), most studies focus on groups A, B1, B2 and D.Citation5 Of these, the former 2 mostly comprise commensal strains, while the latter 2 comprise mostly virulent strains. These belong to a number of major clones represented by multi-locus sequence types (MLST) ST131, ST73, ST95, ST127 (all B2) and ST69 (D).Citation6,7 ExPEC have also been defined by the carriage of various virulence genes encoding adhesins (pap, fimH, sfa/foc), siderophores (fyuA, iutA), toxins (hlyA, cnf1), capsules (kpsMTII) and miscellaneous proteins (traT, ompT, usp and malX), which enable ExPEC to colonise and invade the urinary tract and bloodstream.Citation8-10 Antibiotic resistance is commonly associated with some of these clones, further facilitating their success by evasion of antimicrobial therapy. Notably isolates belonging to ST69 are linked with high rates of trimethoprim-sulfamethoxazole resistance and the ST131-O25b clone is associated with both fluoroquinolone and cephalosporin resistance (CTX-M-15).Citation11,12
Given the problems in the treatment and control of ExPEC infections there is a clear need to understand the factors contributing to their success and dissemination. A greater understanding of the relative importance of virulence determinants versus strain type should help in directing the development of novel drug treatments or ExPEC targeted vaccines.
The greater wax moth caterpillar, Galleria mellonella, has been used in a number of studies as an effective model for investigating the pathogenesis, virulence and antibiotic treatment of many bacterial and fungal pathogens.Citation13-16 Advantages of this model include the ability to incubate larvae over a range of temperatures (15–37°C), adaption to low- and high-throughput experiments, relative ease of maintenance and handling and that the bacteria/drug inoculum can be accurately quantified prior to infection.Citation17-19 In addition to assessing crude mortality, both innate immune responses and cellular damage can easily be monitored post infection.Citation20 Melanin production, induced by G. mellonella during the acute phase response and analogous to the human complement cascade, can be easily assayed.Citation21-23 While, lactate dehydrogenase (LDH) released from cells during apoptosis can be used as a marker of cellular damage.Citation24,25 Therefore, both melanin and LDH production can be used as quantifiable indicators of immunogenicity and response.
G. mellonella has already been validated as a model to investigate the virulence of ExPEC strains from bloodstream infections. This study established that live bacteria with high aggregate virulence scores are required for infection in this model, but the relative importance of other ExPEC variables (phylogroup, sequence type) has not been fully assessed.Citation26 In this work we analyzed 40 ExPEC strains isolated from patients with UTI and/or bloodstream infections encompassing all 4 phylogroups and the clinically relevant major sequence types: ST131, ST127, ST95, ST73 and ST69. We sought to identify ExPEC-specific determinants associated with either heightened mortality and/or a strong immune response in the ExPEC/G. mellonella model.
Results
Galleria mellonella killing
Details of the 40 clinical ExPEC strains studied are summarised in . When infected with 105 CFU/larvae, killing of G. mellonella varied markedly among ExPEC strains, ranging from 0% to 100% (), compared to the CFT073 control strain which affected 100% mortality. Survival curves were analyzed according to patient and strain variables and the relative risk (RR) of each parameter in the Galleria model calculated (). No individual factor could be linked to heightened mortality, although strains from complicated urinary tract infections and/or episodes of urosepsis appeared to be more virulent, compared to those recovered from individuals with asymptomatic bacteruria, cystitis or bacteraemia with a non-urinary focus (p < 0.0001) ().
Table 1. Patient and strain characteristics for 40 extra-intestinal pathogenic Escherichia coli strains. NB: U (urine), B (bacteraemia), ABU (asymptomatic bacteruria), UC (uncomplicated cystitis), COMP (complicated cystitis/pyelonephritis), Chest (respiratory tract source), GU (genitourinary source), GIT (gastrointestinal tract source), Line (intravenous line source), SSTI (skin and soft tissue source), CSF (cerebral spinal fluid source), HC (healthcare setting), CA (community-associated), HA (hospital-associated), F (female), M (male), ST (sequence types), NK (unknown), NT (non-typeable). (*) corresponds to urosepsis patients; (−) indicates not investigated.
Table 2. Percentage mortality and relative risk associated with patient and ExPEC variables Percentage prevalence or mortality is listed with the number of isolates in parenthesis. Compared to the PBS control all variables were significantly associated with increased mortality (P < 0.01), except for patient sex, respiratory-source bacteraemia, ST127, G-fimbriae, CS31A antigen, EAST1 toxin and the K5 antigen (P>0.05). NB: CAI (community-associated infection), HAI (hospital-associated infection), UTI (urinary tract infection), ABU (asymptomatic bacteruria), UC (uncomplicated cystitis), COMP (complicated cystitis/pyelonephritis), GU (genitourinary source), GIT (gastrointestinal source), Line (intravenous line source), Chest (respiratory source), CSF (cerebral spinal fluid source) and SSTI (skin and soft tissue infection source).
Figure 1. Percentage larvae survival, melanin production and LDH production for each of the 40 clinical ExPEC strains. Mean melanin production (blue dots) is plotted on the left axis and mean LDH production (green dots) is plotted on the right axis, against the percentage larvae survival (96-hours post-inoculation, pooled from all 3 killing assays) plotted on the x-axis. Linear regression revealed melanin production to be inversely related to larvae survival (P < 0.0001) only.
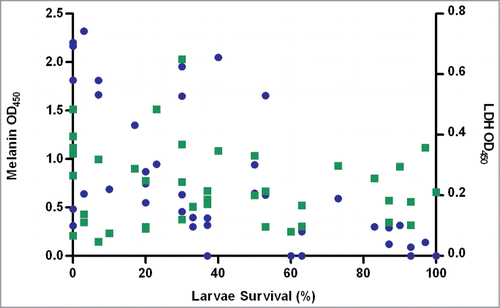
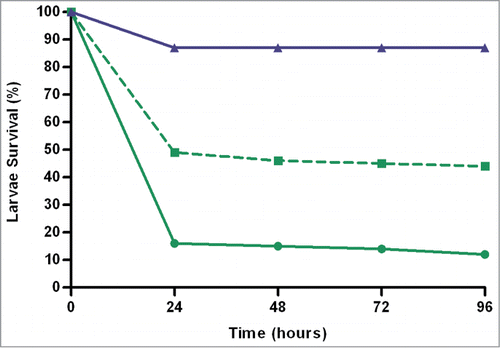
Figure 2. Larvae survival of ExPEC strains associated with urosepsis versus non-urosepsis syndromes. Percentage survival was calculated from 3 separate killing assays 96-hours post-inoculation. Data for strains from urosepsis (n = 6) and other infection types (n = 34) are represented by solid and dashed green lines, respectively. The PBS control is represented in blue. Urosepsis strains affected significantly lower survival (16.2%, P < 0.01) than strains from patients with a UTI only or non-urinary source of bacteraemia (47.2%).
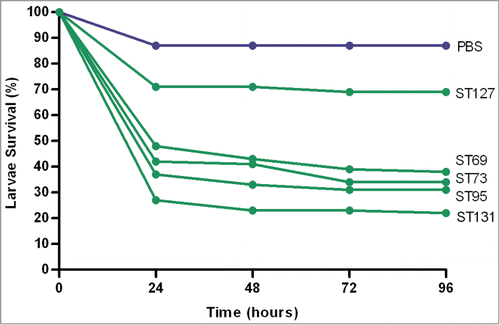
Across the main phylogroups, strains belonging to phylogroup A were more virulent than those belonging to groups B1, B2 and D. Among the major sequence types ST131 was significantly more virulent (P < 0.01), closely followed by ST95, while ST127 was the least virulent (). There was little difference in mortality between strains from male vs. female patients, although community-associated infections were associated with a faster rate of killing compared to hospital-associated strains.
Figure 3. Comparative virulence of the major ExPEC sequence types. Percentage survival was calculated from 3 separate killing assays 96-hours post-inoculation. ST131 isolates (n = 4) to affected the lowest survival (22%, RR 5.88), closely followed by ST95 (n = 4, 31%, RR 5.19), ST73 (n = 3, 34%, RR 4.42) and ST69 (n = 3, 38%, RR 4.67), with ST127 isolates (n = 4) affecting the highest survival (69%, RR 2.31). The PBS control (blue line) is also included for comparison.
Galleria mellonella immune response
Mean melanin and LDH production readings were 0.81 ± 0.11 and 0.23 ± 0.02, respectively. Melanin production varied between strains () and was inversely related with mortality. For example, strains 13, 15, 17, 31, 35 and 38 exhibited the highest percentage survival (>80%) and lowest levels of melanin production (0–0.32). Lower melanin production was significantly associated with a gastrointestinal-source of bloodstream infection (P=0.004). There was no correlation between the production of LDH and overall mortality ().
Galleria mellonella and ExPEC virulence factors
Mortality and relative risk ratios (RR) associated with individual virulence factors are also listed in . Virulence factors (VF) found to be independently linked with mortality (P < 0.001) included afa/dra (97% mortality, RR 7.25) and ompT (87% mortality, RR 6.5). However, these VFs were found in low abundance in these ExPEC strains (≤10%). The most prevalent ExPEC VFs were fimH (75%), fyuA (75%), usp (58%), traT (53%), pap, kpsII and malX (all 50%). Of these, pap was associated with higher mortality than fimH; fyuA exhibited the lowest mortality among all of the siderophores; isolates possessing kpsII were similar to kpsIII, although strains with K1 were linked to significantly higher mortality; while traT, usp and malX shared similar RR, but mortality was higher with strains possessing traT. Of the 2 serogroups analyzed in this study, the O6-antigen was associated with significantly higher larvae killing than the O25-antigen (93%, RR 7.00 vs. 78%, RR 5.88).
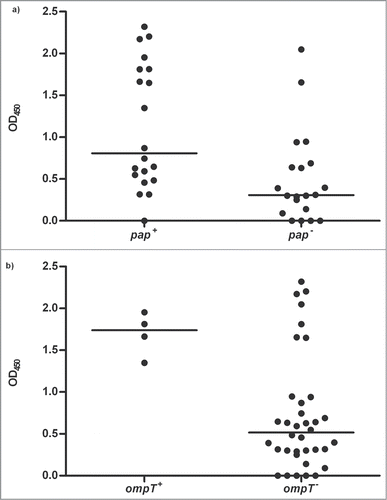
Melanin production was highest for isolates possessing pap (1.13), cnf1 (1.15), ompT (1.70) and sat (1.10). However, the Mann-Whitney test revealed the majority of pap-positive (P < 0.01) and ompT-positive (P < 0.01) strains induced significantly higher melanin production compared to strains lacking these VFs, despite a similar spread in OD readings for strains possessing and lacking these structures ().
Figure 4. Melanin production induced by strains possessing pap (n = 20) or ompT (n = 4) vs. strains lacking these virulence factors. The + and – indicates presence and absence of pap (panel a) and ompT (panel B); each dot represents the median melanin OD 4-hours post-inoculation for one isolate. The Mann-Whitney test revealed the presence of both pap (P = 0.004) and ompT (P = 0.02) to be associated with increased melanin production.
Although VF and Abx scores differed markedly among isolates, linear regression demonstrated no relationship between either of these parameters and mortality, melanin or LDH production in G. mellonella.
Discussion
The G. mellonella infection model has been used to investigate the virulence of a range of clinically relevant bacterial and fungal pathogens, but was only recently established as a viable model for analyzing ExPEC virulence.Citation16,19,20,26,27 Williamson et al (2014) analyzed 9 E. coli strains recovered from bloodstream infections and found a correlation between a high aggregate virulence score and larvae mortality.Citation26 The ExPEC population, however, is highly heterogeneous, containing strains of multiple phylogroups, sequence types, serogroups and virulence factor profiles.Citation6,28 We therefore sought to analyze a wider array of clinical ExPEC strains encompassing all of these variables using the G. mellonella model.
The heterogeneity of these clinical ExPEC strains and varied survival indicates that virulence may be clonal in nature, as reported for K. pneumoniae clinical strains by Wand et al (2013).Citation25 Analysis of K. pneumoniae strains also revealed LDH to be the best marker of virulence in the G. mellonella model, but this was not observed for ExPEC strains here. Using LDH as a marker, it is difficult to determine whether this cell damage is due to specific or multiple virulence factors or an overwhelming immune response to infection. In addition, it is unclear how well optical density corresponds with cell damage and the quantity of LDH released. Bacterial quantification in the hemolymph, versus other larvae structures, may provide a more accurate measure of cell damage in future studies.
Retrospective clinical studies demonstrate that E. coli bacteraemia is associated with a mortality rate between 17% and 61%.Citation29,30 The incidence is increasing each year in the UK (www.hpa.org.uk), with the urinary tract identified as the most common source.Citation1 This trend toward increased virulence among E. coli urosepsis strains is reflected in G. mellonella whereby, larvae mortality was found to be significantly higher in strains associated with urosepsis compared to those from patients with an uncomplicated UTI or from a non-urinary source of bacteraemia. In addition, isolates belonging to ST131 affected the highest mortality suggesting that they may be significantly more virulent than the other major lineages. ST131 is frequently detected in urinary and bloodstream infections, therefore, this finding is not surprising.Citation1,31 However, comparison of strains possessing the O6 antigen vs. those possessing the O25 antigen revealed higher mortality was associated with O6 (93%, RR 7.00 vs. 78%, RR 5.88). Given the lower mortality observed with isolates belonging to ST73 and, especially, 3 of the 4 ST127 strains, it could be concluded that O25 strains are more virulent. A recent study has shed light on this phenomenon, whereby an insertion sequence in the O6-antigen of ST73 and ST127 strains was associated with heightened virulence, but those lacking this sequence were avirulent.Citation32 This may explain the low mortality seen with the majority of our ST127 strains, despite them belonging to phylogroup B2.
Despite the array of virulence factors possessed by ExPEC strains, only a few individual determinants were associated with heightened mortality and/or a strong immunological response. AFA/Dr adhesins and OmpT were associated with significant mortality, although their genes were only carried by 10% of strains. OmpT and P-fimbriae possessing strains generated significantly higher melanin compared to strains lacking these surface structures. Outer membrane proteins of K. pneumoniae, such as OmpA, have also been shown to contribute to virulence in G. mellonella and P-fimbriae have been linked to adverse patient outcomes in clinical studies.Citation20,33 These VFs may well have significant roles in urosepsis caused by ExPEC strains, as 3 of the 4 ST131 strains analyzed here possessed pap, ompT and/or afa/dra, but a larger study of these factors would need to be performed to confirm these findings.
Next generation sequencing has proven invaluable as a tool for understanding the complexity of E. coli and their different pathotypes.Citation34,35 Use of this technology to identify the clonal features, such as genomic fragments, associated with virulence and mortality, in conjunction with a larger study of ExPEC lineages, would help validate these findings in E. coli.
Williamson et al (2014) demonstrated that the G. mellonella model was comparable to a mouse model for investigating the virulence of clinical ExPEC strains.Citation26 In this study we have used the model to identify ST131, a genitourinary-source of bacteraemia and pap and ompT as important variables in the pathogenicity of ExPEC. Future studies characterizing ExPEC strains from fatal cases of E. coli urosepsis will be used to confirm these observations and also identify new variables associated with patient mortality, bringing together the clinical and microbiological aspects of E. coli bacteraemia to improve future patient management.
Materials and Methods
Characterization of Escherichia coli Isolates
Forty characterized ExPEC isolates were selected from 2 large collections of E. coli clinical isolates that have been described previously elsewhere.Citation36 MICs to ampicillin, amoxicillin-clavulanate, aztreonam, ceftazidime, cefotaxime, cefpodoxime, cefoxitin, piperacillin-tazobactam, meropenem, ciprofloxacin, tobramycin, gentamicin, minocycline, temocillin, tigecycline, chloramphenicol, trimethoprim-sulfamethoxazole and nitrofurantoin had been determined by agar dilution, according to BSAC breakpoints.Citation37 Isolates recovered from patients diagnosed with either an upper UTI accompanied by systemic symptoms or with a bloodstream infection arising from a urinary-source was classified as urosepsis strains.
Thirty-six common ExPEC virulence factors were also identified using an updated (Personal communication with Prof. James Johnson, University of Minnesota, 2012) PCR assay, consisting of 6 multiplex PCRs. Thirty-six different virulence factors were detected, ranging in prevalence from 5% (afa/dra) to 75% (fimH and fyuA). Virulence factors detected are listed in . Each isolate was given a virulence factor (VF) score, calculated as the number of virulence factors detected.Citation38 Multiple PCR targets for the same operon, such as pap, were scored as one. Each isolate was also given an antimicrobial resistance (Abx) score, calculated as the sum of antibiotics to which the isolate demonstrated resistance. Strain characteristics are listed in .
Galleria mellonella Killing Assay
G. mellonella final instar larvae were obtained from Livefoods UK Ltd and stored on wood chips in the dark at 15°C, for up to one week, until use. Prior to and during all experiments, food was provided in the form of wood chips. The optimal inoculum for the killing assays was determined by injecting 10 larvae with 104, 105 and 106 CFU/larvae and identifying the inoculum which killed 50% of larvae after 24 hours incubation at 37°C.
The optimal bacterial inoculum (105CFU/larvae) and injections were prepared using published methods.Citation19 Each killing assay included 10 inoculated larvae per ExPEC isolate. Mock inoculated (PBS) larvae were included to control for any lethal effects of the injection process and the CFT073 E. coli pyelonephritis type strain was used as a positive control.Citation39 Larvae were incubated for 96 hours at 37°C, in the dark, and scored every 24 hours for live and dead larvae. Larvae which failed to respond to touch were recorded as dead. All killing assays were performed in triplicate, over 3 separate weeks, using 3 different batches of G. mellonella larvae.
Galleria mellonella melanisation assay
Melanin production was measured as described by Wand et al.Citation25 Larvae were inoculated as described in the killing assay. Larvae were incubated for 4 hours at 37°C in the dark and then chilled on ice for 5 minutes. Larvae were surfaced sterilised with 70% ethanol (Thermo Fisher Scientific), sacrificed using a sterile disposable surgical scalpel (Swann-Morton) and hemolymph collected into a pre-chilled eppendorf tube containing 10–20 crystals of N-phenylthiourea (Alfa Aesar) to prevent further melanisation. Citation40 Optical density (OD) was measured at 450nm using an ELx800 plate reader (Biotek).
E. coli CFT073- and PBS-inoculated larvae were used as positive and negative controls, respectively. The OD of hemolymph collected from uninfected larvae was subtracted from all test readings to correct for the background. All experiments were performed in triplicate, as described above.
Galleria mellonella lactate dehydrogenase assay
This assay was performed as described by Wand et al., using the CytoTox 96 Non-radioactive Cytotoxicity Assay (Promega), according to the manufacturer's instructions.Citation25 Hemolymph was collected as described for the melanisation assay. OD readings and interpretation was performed as for the melanisation assay. All experiments were performed in triplicate, as described above.
Statistical analysis
Percentage larvae survival corresponded to the total number of larvae that survived, 96-hours post-inoculation, from the 3 pooled experiments. Mean melanin and LDH production corresponded to the OD, 4-hours post-inoculation, from the 3 pooled experiments. All statistical analyses were performed using GraphPad Prism version 5.04.
Survival analysis: all 3 independent sets of survival data were pooled for analysis, for accurate comparison with the PBS (negative) control and CFT073 (positive) control. The total proportion of larvae that died, for each patient and strain variable was compared to the PBS control to determine mortality and a relative risk ratio (RR) generated to identify the variables associated with an increased risk of death. Association between percentage survival and VF score and Abx score was measured by linear regression. A P-value ≤0.05 indicated significance.
Melanin/LDH analysis: association between survival and melanin production or LDH production was measured by linear regression. A P-value ≤0.01 indicated statistical significance for both assays.
Differences in melanin or LDH production, in relation to patient or strain variables, were measured using the Mann-Whitney test. A P-value ≤0.05 indicated statistical significance.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
988095_Supplementary_Materials.zip
Download Zip (16.1 KB)Acknowledgments
We are grateful to Dr Alan McNally (Nottingham Trent University) for providing the CFT073 type strain.
funding
This work was supported by the Public Health England PhD studentship fund.
References
- Horner C, Fawley W, Morris K, Parnell P, Denton M, Wilcox M. Escherichia coli bacteraemia: 2 years of prospective regional surveillance (2010-12). J Antimicrob Chemother 2014; 69(1):91-100; PMID:24003184; http://dx.doi.org/10.1093/jac/dkt333
- Gibreel T, Dodgson A, Cheesbrough J, Fox A, Bolton F, Upton M. Population structure, virulence potential and antibiotic susceptibility of uropathogenic Escherichia coli from Northwest England. J Antimicrob Chemother 2012; 67(2):346-356; PMID:22028202; http://dx.doi.org/10.1093/jac/dkr451
- Abdallah KS, Cao Y, Wei DJ. Epidemiologic Investigation of extra-intestinal pathogenic E. coli (ExPEC) based on PCR phylogenetic group and fimH single nucleotide polymorphisms (SNPs) in China. Int J Mol Epidemiol Genet 2011; 2(4):339-353; PMID:22199997
- Russo T, Johnson J. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J Infect Dis 2000; 181(5):1753-1754; PMID:10823778; http://dx.doi.org/10.1086/315418
- Clermont O, Christenson JK, Denamur E, Gordon DM. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep 2013; 5(1):58-65; PMID:23757131; http://dx.doi.org/10.1111/1758-2229.12019
- Banerjee R, Johnston B, Lohse C, Chattopadhyay S, Tchesnokova V, Sokurenko E, Johnson JR. The clonal distribution and diversity of extraintestinal Escherichia coli varies according to patient characteristics. Antimicrob Agents Chemother 2013; 57(12):5912-5917; PMID:24041881; http://dx.doi.org/10.1128/AAC.01065-13
- Lau S, Reddy S, Cheesbrough J, Bolton F, Willshaw G, Cheasty T, Fox A, Upton M. Major uropathogenic Escherichia coli strain Isolated in the North West of England identified by multilocus sequence typing. J Clin Microbiol 2008; 46(3):1076-1080; PMID:18199778; http://dx.doi.org/10.1128/JCM.02065-07
- Johnson JR, Russo TA. Molecular epidemiology of extraintestinal pathogenic (uropathogenic) Escherichia coli. Int J Med Microbiol 2005; 295(6-7):383-404; PMID:16238015; http://dx.doi.org/10.1016/j.ijmm.2005.07.005
- Cooke N, Smith S, Kelleher M, Rogers T. Major differences exist in frequencies of virulence factors and multidrug resistance between community and nosocomial Escherichia coli bloodstream isolates. J Clin Microbiol 2010; 48(4):1099-1104; PMID:20107091; http://dx.doi.org/10.1128/JCM.02017-09
- Pitout JD, Laupland KB, Church DL, Menard ML, Johnson JR. Virulence factors of Escherichia coli isolates that produce CTX-M-type extended-spectrum β-lactamases. Antimicrob Agents Chemother 2005; 49(11):4667-4670; PMID:16251310; http://dx.doi.org/10.1128/AAC.49.11.4667-4670.2005
- Johnson J, Menard M, Lauderdale T, Kosmidis C, Gordon D, Collignon P, Maslow J, Andrasevic A, Kuskowski M, Trans-global initiative for antimicrobial resistance analysis investigators. Global distribution and epidemiologic associations of Escherichia coli clonal group A, 1998-2007. Emerg Infect Dis 2011; 17(11):2001-2009; PMID:22099087; http://dx.doi.org/10.3201/eid1711.110488
- Nicolas-Chanoine M, Blanco J, Leflon-Guibout V, Demarty R, Alonso M, Canica M, Park Y, Lavigne J, Pitout J, Johnson J. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J Antimicrob Chemother 2008; 61(2):273-281; PMID:18077311; http://dx.doi.org/10.1093/jac/dkm464
- Glavis-Bloom J, Muhammed M, Mylonakis E. Of model hosts and man: using Caenorhabditis elegans, Drosophila melanogaster and Galleria mellonella as model hosts for infectious disease research. Adv Exp Med Biol 2012; 710:11-17; PMID:22127881; http://dx.doi.org/10.1007/978-1-4419-5638-5_2
- Joyce SA, Gahan CG. Molecular pathogenesis of Listeria monocytogenes in the alternative model host Galleria mellonella. Microbiology 2010; 156(Pt 11):3456-3468; PMID:20688820; http://dx.doi.org/10.1099/mic.0.040782-0
- Miyata S, Casey M, Frank DW, Ausubel FM, Drenkard E. Use of the Galleria mellonella caterpillar as a model host to study the role of the type III secretion system in Pseudomonas aeruginosa pathogenesis. Infect Immun 2003; 71(5):2404-2413; PMID:12704110; http://dx.doi.org/10.1128/IAI.71.5.2404-2413.2003
- Hornsey M, Phee L, Longshaw C, Wareham DW. In vivo efficacy of telavancin/colistin combination therapy in a Galleria mellonella model of Acinetobacter baumannii infection. Int J Antimicrob Agents 2013; 41(3):285-287; PMID:23312607; http://dx.doi.org/10.1016/j.ijantimicag.2012.11.013
- Aperis G, Fuchs BB, Anderson CA, Warner JE, Calderwood SB, Mylonakis E. Galleria mellonella as a model host to study infection by the Francisella tularensis live vaccine strain. Microbes Infect 2007; 9(6):729-734; PMID:17400503; http://dx.doi.org/10.1016/j.micinf.2007.02.016
- Nicoletti M, Iacobino A, Prosseda G, Fiscarelli E, Zarrilli R, De Carolis E, Petrucca A, Nencioni L, Colonna B, Casalino M. Stenotrophomonas maltophilia strains from cystic fibrosis patients: genomic variability and molecular characterization of some virulence determinants. Int J Med Microbiol 2011; 301(1):34-43; PMID:20952251; http://dx.doi.org/10.1016/j.ijmm.2010.07.003
- Betts JW, Phee LM, Woodford N, Wareham DW. Activity of colistin in combination with tigecycline or rifampicin against multidrug-resistant Stenotrophomonas maltophilia. Eur J Clin Microbiol Infect Dis 2014; PMID:24781003
- Insua JL, Llobet E, Moranta D, Perez-Gutierrez C, Tomas A, Garmendia J, Bengoechea JA. Modeling Klebsiella pneumoniae pathogenesis by infection of the wax moth Galleria mellonella. Infect Immun 2013; 81(10):3552-3565; PMID:23836821; http://dx.doi.org/10.1128/IAI.00391-13
- Nappi AJ, Christensen BM. Melanogenesis and associated cytotoxic reactions: applications to insect innate immunity. Insect Biochem Mol Biol 2005; 35(5):443-459; PMID:15804578; http://dx.doi.org/10.1016/j.ibmb.2005.01.014
- Lee YS, Yun EK, Jang WS, Kim L, Lee JH, Park SY, Ryu KS, Seo SJ, Kim CH, Lee IH. Purification, cDNA cloning and expression of an insect defensin from the great wax moth, Galleria mellonella. Insect Mol Biol 2004; 13(1):65-72; PMID:14728668; http://dx.doi.org/10.1111/j.1365-2583.2004.00462.x
- Lionakis MS. Drosophila and Galleria insect model hosts: new tools for the study of fungal virulence, pharmacology and immunology. Virulence 2011; 2(6):521-527; PMID:22186764; http://dx.doi.org/10.4161/viru.2.6.18520
- Berg J, Tymoczko J, Stryer L. Biochemistry. 5th ed. New York: W H Freeman, 2002.
- Wand M, McCowen J, Nugent P, Sutton J. Complex interactions of Klebsiella pneumoniae with the host immune system in a Galleria mellonella infection model. J Med Microbiol 2013; 62(Pt 12):1790-1798; PMID:24000226; http://dx.doi.org/10.1099/jmm.0.063032-0
- Williamson DA, Mills G, Johnson JR, Porter S, Wiles S. In vivo correlates of molecularly inferred virulence among extraintestinal pathogenic Escherichia coli (ExPEC) in the wax moth Galleria mellonella model system. Virulence 2014; 5(3):388-393; PMID:24518442; http://dx.doi.org/10.4161/viru.27912
- Mylonakis E, Moreno R, El Khoury JB, Idnurm A, Heitman J, Calderwood SB, Ausubel FM, Diener A. Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect Immun 2005; 73(7):3842-3850; PMID:15972469; http://dx.doi.org/10.1128/IAI.73.7.3842-3850.2005
- Peirano G, Mulvey GL, Armstrong GD, Pitout JD. Virulence potential and adherence properties of Escherichia coli that produce CTX-M and NDM β-lactamases. J Med Microbiol 2013; 62(Pt 4):525-530; PMID:23319311; http://dx.doi.org/10.1099/jmm.0.048983-0
- Schlackow I, Stoesser N, Walker AS, Crook DW, Peto TE, Wyllie DH. Increasing incidence of Escherichia coli bacteraemia is driven by an increase in antibiotic-resistant isolates: electronic database study in Oxfordshire 1999-2011. J Antimicrob Chemother 2012; 67(6):1514-1524; PMID:22438437; http://dx.doi.org/10.1093/jac/dks082
- Melzer M, Petersen I. Mortality following bacteraemic infection caused by extended spectrum β-lactamase (ESBL) producing E. coli compared to non-ESBL producing E. coli. J Infect 2007; 55(3):254-259; PMID:17574678; http://dx.doi.org/10.1016/j.jinf.2007.04.007
- Colpan A, Johnston B, Porter S, Clabots C, Anway R, Thao L, Kuskowski MA, Tchesnokova V, Sokurenko EV, Johnson JR. Escherichia coli sequence type 131 (ST131) sub-clone H30 as an emergent multidrug-resistant pathogen among US veterans. Clin Infect Dis 2013; 57(9):1256-1265; PMID:23926176; http://dx.doi.org/10.1093/cid/cit503
- Alghoribi MF, Gibreel TM, Dodgson AR, Beatson SA, Upton M. Galleria mellonella Infection Model Demonstrates High Lethality of ST69 and ST127 Uropathogenic E. coli. PLoS One 2014; 9(7):e101547; PMID:25061819; http://dx.doi.org/10.1371/journal.pone.0101547
- Mora-Rillo M, Fernández-Romero N, Navarro-San Francisco C, Romero-Gómez MP, Pano-Pardo JR, Arribas-López JR, Mingorance J. The presence of the fyuA and P fimbriae genes are related with mortality in Escherichia coli bloodstream infections. 23rd European Congress of Clinical Microbiology and Infectious Disease; 2013 Apr 27-30; Berlin, Germany.
- McNally A, Alhashash F, Collins M, Alqasim A, Paszckiewicz K, Weston V, Diggle M. Genomic analysis of extra-intestinal pathogenic Escherichia coli urosepsis. Clin Microbiol Infect 2013; 19(8):E328-334; PMID:23573792; http://dx.doi.org/10.1111/1469-0691.12202
- Underwood AP, Dallman T, Thomson NR, Williams M, Harker K, Perry N, Adak B, Willshaw G, Cheasty T, Greeen J. et al. Public health value of next-generation DNA sequencing of enterohemorrhagic Escherichia coli isolates from an outbreak. J Clin Microbiol 2013; 51(1):232-237; PMID:23135946; http://dx.doi.org/10.1128/JCM.01696-12
- Ciesielczuk H, Hornsey M, Choi V, Woodford N, Wareham D. Development and evaluation of a multiplex PCR for eight plasmid-mediated quinolone resistance determinants. J Med Microbiol 2013; 62(Pt 12):1823-1827; PMID:24000223; http://dx.doi.org/10.1099/jmm.0.064428-0
- Andrews JM. Determination of minimum inhibitory concentrations. J Antimicrob Chemother 2001; 48 Suppl 1:5-16; http://dx.doi.org/10.1093/jac/48.suppl_1.5
- Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis 2000; 181(1):261-272; http://dx.doi.org/10.1086/315217
- Mobley HL, Green DM, Trifillis AL, Johnson DE, Chippendale GR, Lockatell CV, Jones BD, Warren JW. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect Immun 1990; 58(5):1281-1289; PMID:2182540
- Andrejko M, Mizerska-Dudka M. Effect of Pseudomonas aeruginosa elastase B on level and activity of immune proteins/peptides of Galleria mellonella hemolymph. J Insect Sci 2012; 12:88; PMID:23421724; http://dx.doi.org/10.1673/031.012.8801
