GFP-Atg8/LC3 is a very useful marker to follow macroautophagy (hereafter autophagy). In contrast, Atg8/LC3-GFP has relatively limited uses. Unfortunately, many authors do not seem to appreciate the difference between these two constructs. I hope to put an end to that now.
The title of this missive pretty much makes my point, but I will elaborate briefly. Atg8/LC3 is of course a key autophagy-related protein (for the purposes of this article I am going to ignore the other LC3 family members, but the same holds true for them). Although its function is not clear, we do know that it is a ubiquitin-like protein that undergoes conjugation to phosphatidylethanolamine (PE). Importantly, Atg8/LC3 remains associated with the completed autophagosome, and for this reason it is the key autophagy-related marker protein; when fused to GFP or similar fluorophores it can be used to follow the phagophore or autophagosome in vivo.
Let’s quickly review what is known about the conjugation process. In most organisms Atg8/LC3 is initially synthesized with a C-terminal amino acid(s) that follow the critical glycine residue. The additional amino acid (or amino acids) is removed by the Atg4 cysteine protease, exposing the glycine residue. Subsequent activation and conjugation involve Atg7 and Atg3, respectively ().
Figure 1. Schematic comparison of the fate of C-terminal versus N-terminal GFP fused to Atg8. (A) If GFP is fused to the C terminus of Atg8 (or LC3), it is removed when Atg4 hydrolyzes the bond between the penultimate glycine residue and arginine. The result is free GFP (with an additional arginine) that does not serve as a marker for the phagophore or autophagosome. However, a C-terminal fusion can be used to monitor Atg4 activity. (B) If GFP is fused to the N terminus of Atg8 (LC3), it remains attached to the protein until vacuolar (lysosomal) delivery; Atg8 is degraded relatively rapidly compared to GFP, so the generation of free GFP in the vacuolar/lysosomal lumen can be used to monitor autophagy activity. Furthermore, the stable GFP-Atg8/LC3 chimera can be used to follow the phagophore or autophagosome. Although the schematic refers to yeast Atg8, essentially the same hold true for LC3 and its related family members.
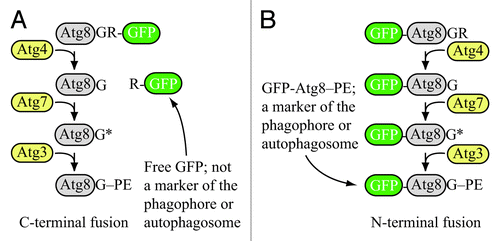
Now, let’s return to that first step, Atg4-dependent cleavage of the C terminus. You see, that is my point. Atg4 cleaves the C terminus of Atg8/LC3 after the glycine residue. To put it another way, anything after that glycine will be removed from Atg8/LC3 early on in the process of autophagy. Got it? Removed. Gone. No longer connected to Atg8/LC3.
In yeast, a C-terminal fusion of GFP to Atg8 (written as Atg8-GFP) has been used to monitor Atg4 activity (). Otherwise, the fusion is always at the N terminus. With an N-terminal fusion, GFP-Atg8/LC3 can be used as a marker of the phagophore or the autophagosome as noted above. A tandem mCherry-GFP-LC3 (or similar construct) can be used to monitor flux, or at least the conversion of autophagosomes to autolysosomes. Similarly, GFP-Atg8/LC3 can be followed by western blot, and the appearance of free GFP is indicative of autophagic flux ().
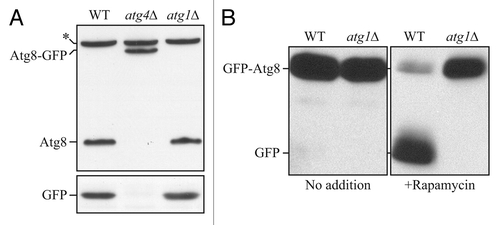
Figure 2. Atg8-GFP cannot be used to monitor autophagic flux. (A) Wild-type (WT), atg4Δ or atg1Δ cells expressing Atg8-GFP were grown to mid-log phase and protein extracts were examined by western blot with antisera to Atg8 or GFP. The asterisk indicates a nonspecific band. With a C-terminal fusion, GFP is cleaved from Atg8 without shifting to autophagy-inducing conditions, and even in an autophagy-defective mutant; however, the Atg8-GFP construct can be used to monitor Atg4 activity. This figure was modified from data previously published in reference Citation1, and is reproduced by permission of the American Society for Cell Biology, copyright 2001. (B) WT or atg1Δ cells expressing GFP-Atg8 were grown to mid-log phase and divided into aliquots; one set of samples was treated with rapamycin and protein extracts were examined by western blot with antiserum to GFP. In this case, free GFP is not generated prior to autophagy induction or in an autophagy-defective mutant. Thus, the N-terminal fusion of GFP can be used to monitor autophagy induction, and also serves as a marker for the phagophore and autophagosome (as well as Cvt or autophagic bodies if breakdown is prevented in the vacuolar lumen, for example, by the addition of PMSF or the use of a mutant such as pep4Δ). This figure was modified from data previously published in reference Citation2, and is reproduced by permission of the American Society for Cell Biology, copyright 2007.
The point is, if you want to use a fluorescently tagged Atg8/LC3 to monitor anything other than Atg4 activity, you have to use an N-terminal fusion (written as GFP-Atg8/LC3). Because of these two potential uses for Atg8/LC3, it is important to specify the construct correctly. Furthermore, the incorrect written usage of Atg8/LC3-GFP indicates a fundamental lack of understanding of the conjugation process (or simply carelessness). Nonetheless, I still see authors incorrectly referring to Atg8/LC3-GFP. For example, looking at abstracts from a few papers published in 2009 and 2010, I found the following (if I extend my search beyond the abstracts I could provide many more examples, including papers published in 2011):
“...L. monocytogenes was encapsulated by LC3-GFP...”
“...NRK-52E cell lines stably transfected with LC3-GFP...”
“...large vesicles that intensely stained for a transfected LC3-GFP construct...”
“...LC3-GFP fusion protein was accumulated as granular dots in autophagosomes.”
“An autophagosome marker protein Atg8-GFP...”
I highly doubt the technical validity of these statements. For example, if Atg4 does not remove GFP from the C terminus, there is no way the fusion protein is going to get conjugated to PE. Similarly, I find it unlikely that the authors used Atg8/LC3-GFP, because they wanted to monitor autophagosome formation, not Atg4 activity. I could give you more examples, but I think you get the point.
So, unless you are talking about an assay for Atg4, please use the correct designation, GFP-Atg8/LC3.
References
- Kim J, Huang W-P, Klionsky DJ. Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J Cell Biol 2001; 152:51 - 64; http://dx.doi.org/10.1083/jcb.152.1.51; PMID: 11149920
- Yorimitsu T, Zaman S, Broach JR, Klionsky DJ. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae.. Mol Biol Cell 2007; 18:4180 - 9; http://dx.doi.org/10.1091/mbc.E07-05-0485; PMID: 17699586