Abstract
Many proteins involved in autophagy have been identified in the yeast Saccharomyces cerevisiae. For example, Atg3 and Atg10 are two E2 enzymes that facilitate the conjugation of the ubiquitin-like proteins (Ubls) Atg8 and Atg12, respectively. Here, we describe the identification and characterization of the predicted Atg10 homolog (SpAtg10) of the evolutionarily distant Schizosaccharomyces pombe. Unexpectedly, SpAtg10 is not essential for autophagy. Instead, we find that SpAtg10 is essential for normal cell cycle progression, and for responses to various stress conditions that perturb the cell cycle, independently of Atg12 conjugation. Taken together, our data indicate that autophagic Ubl conjugation pathways differ between eukaryotes and, furthermore, that enzymes such as Atg10 may have additional functions in controlling key cellular processes such as cell cycle progression. Atg10-related proteins are found from yeast to humans, and, thus, this study has implications for understanding the functions of this protein family in Ubl conjugation in eukaryotes.
Introduction
Ubiquitin (Ub) and ubiquitin-like proteins (Ubls) such as SUMO are reversibly conjugated to proteins by very similar enzymatic pathways.Citation1,Citation2 First, an activating enzyme (E1) adenylates the Ub/Ubl molecule at the C terminus before forming a thioester bond to the modifier via a catalytic cysteine residue. The Ub/Ubl is then transferred to a conjugating enzyme (E2), again forming a thioester bond to a catalytic cysteine residue. Finally, ligase enzymes (E3s) facilitate Ub/Ubl conjugation to specific substrates, either by binding the modifier directly or by functioning as an adaptor protein. In the majority of cases, the Ub/Ubl forms an isopeptide bond to a lysine residue within the substrate protein. These post-translational modifications play important roles in regulating protein function, for example, by targeting for degradation key regulatory proteins that influence normal cell cycle progression. However, in addition to degradation, Ubl conjugation can influence protein activity, location and protein/protein interactions.Citation2 Interestingly, two different Ubls are conjugated to specific substrates to regulate autophagy in eukaryotes.
In eukaryotes, macroautophagy (hereafter referred to as “autophagy”) is a key process that allows the “self-digestion” of cellular components to maintain homeostasis. Although autophagy appears to function at a basal level in cells, the process can be upregulated in response to various stimuli, including nutrient starvation.Citation3 During autophagy, proteins and organelles are sequestered in a double-membrane vesicle, the autophagosome, and transported to the vacuole/lysosome. The autophagosome fuses with the vacuole/lysosome, and releases its inner vesicle (or “autophagic body”) into the lumen. The autophagic body is dismantled, and its contents degraded by proteolytic enzymes. The molecular control of autophagosome formation has largely been characterized in the budding yeast Saccharomyces cerevisiae and has revealed numerous genes that are required for autophagy in eukaryotes.Citation4-Citation8 Many proteins function in autophagosome formation, including the Ubls Atg12 and Atg8.Citation9 Atg7 acts as the E1 enzyme for Atg12 by activating the Ubl and transferring it to a downstream E2 enzyme, Atg10, which conjugates Atg12 to Atg5.Citation10-Citation12 Atg7 also functions as the E1 enzyme for Atg8 by transferring the Ubl to a different E2 enzyme, Atg3, which then conjugates Atg8 to phosphatidylethanolamine (PE) lipids on the autophagosomal membrane.Citation13-Citation15 Interestingly, the Atg12-Atg5 conjugate functions as a novel E3 ligase to direct Atg8 conjugation to PE.Citation16 A cysteine protease, Atg4, can reverse Atg8 conjugation by removing Atg8 from PE.Citation15 Based on these studies in yeast, and the identification of many homologs of the yeast proteins in other organisms,Citation17 it is widely predicted that autophagic Ubl conjugation pathways are conserved from yeast to human. However, in many cases, it is difficult to identify homologs of autophagic E2 enzymes, particularly Atg10, due to the lack of homology of amino acid sequence across the whole protein. Indeed, a two-hybrid screen was required to identify the Atg10 homolog in mice, which was found to share only 19% homology with its counterpart in budding yeast.Citation18 Importantly, analysis of Atg10 and Atg3 proteins has revealed some limited homology, which is located around the catalytic cysteine residue of these enzymes that is not shared with E2 enzymes in other Ub/Ubl pathwaysCitation11 (). Significantly, although Atg10 is essential for autophagy in S. cerevisiae, the role(s) played by Atg10-like enzymes in other organisms is much less well-defined. Thus, there is still much to learn regarding the function and regulation of the enzymes that participate in Atg12/Atg8 Ubl conjugation pathways.
Figure 1. Bioinformatic analysis of the potential Atg10 homolog in S. pombe. CLUSTAL sequence alignment of Atg10 family members from human (HsAtg10), Caenorhabditis elegans (CeAtg10) and S. cerevisiae (ScAtg10), together with the predicted protein sequence of Spac227.04 (SpAtg10) is shown. Asterisks indicate identical amino acids shared by all the proteins; a colon, a highly similar substitution; full stop, a similar substitution. Bold indicates an amino acid that is found in at least three of the Atg10 proteins and ΨHPC motifs are shown in red.

Here, we identify the only Atg10-like protein (SpAtg10) in the fission yeast Schizosaccharomyces pombe. Unexpectedly, we demonstrate that SpAtg10 is not required for autophagic activity in S. pombe. However, instead, excitingly, we find that SpAtg10, but not Atg12, is required for normal cell cycle progression and for responses to several stress agents that perturb the cell cycle. Although we cannot rule out a role for SpAtg10 in autophagy under all circumstances, our data reveal a hitherto unknown link between Atg10-like proteins and the regulation of the cell cycle in eukaryotes. Thus, taken together, these results have implications for the functions of autophagic E2 enzymes and Ubl conjugation pathways in other organisms.
Results and Discussion
S. pombe contains a single potential Atg10-like protein
The E2 enzyme equivalent to Atg10 in S. cerevisiae and other organisms has not previously been identified in S. pombe.Citation19,Citation20 However, studies in S. cerevisiae demonstrated that Atg10 and Atg3 have similar amino acid sequences at their catalytic sites which are unique to autophagic E2 enzymes,Citation13 and these sequences are also found in Atg10 and Atg3 homologs in other organisms.Citation9 In these autophagic E2 enzymes, the catalytic cysteine residue forms part of an HPC motif, often with a hydrophobic residue located upstream of the histidine residue (hereafter referred to as the ΨHPC motif; ). Hence, to identify the Atg10 homolog in S. pombe, we searched the entire fission yeast-predicted proteome for proteins that contain this ΨHPC motif. Twenty-six of the ~5,000 predicted proteins in S. pombe were found to contain this motif (). As expected, one of these proteins was previously identified as Atg3.Citation19 Of the remaining 25 proteins, 15 were encoded by previously named genes involved in processes unlinked to Ubl modification, and a further seven shared homology with proteins predicted to have roles in other processes (). Of the remaining three proteins (Spbc651.12c, Spbc1271.08c, Spac227.04), the ΨHPC motif was located at the C terminus of Spbc651.12c, and Spbc1271.08c is predicted to contain a transmembrane domain and localizes to the ER.Citation21 The remaining ORF, SPAC227.04, encodes a protein with a predicted molecular weight (Mr) of ~20 kDa, which is the approximate Mr of most E2 enzymes, suggesting that this ORF encodes Atg10. Significantly, and consistent with this hypothesis, analysis of the predicted sequences of Spac227.04 and Atg3 at NCBI revealed that both proteins have significant homology to the previously annotated pfam03987 domain (autophagocytosis-associated protein, active-site domain), as expected for autophagic E2 enzymes. Furthermore, Atg3, but not Spac227.04, also contains significant homology to the previously annotated pfam03986 [autophagocytosis-associated protein (Atg3), N-terminal domain] and pfam10381 (autophagocytosis-associated protein C-terminal), two domains that are predicted to be present in Atg3-like but not Atg10-like E2 enzymes. Finally, ClustalW analysis of the predicted amino acid sequence of Spac227.04 revealed the presence of amino acids that are also found in different, previously identified Atg10 proteins, in addition to the ΨHPC motif (). Thus, taken together, our bioinformatic analyses indicate that SPAC227.04 most likely encodes the Atg10 protein in S. pombe (hereafter referred to as SpAtg10).
Table 1. Open reading frames (ORFs) in S. pombe that contain the ΨHPC motif
SpAtg10 is not involved in autophagy
In contrast to the distantly related S. cerevisiae, the proteins involved in autophagy are relatively uncharacterized in S. pombe. However, recent work revealed that the hydrolysis of an Atg8-GFP fusion protein reflects autophagic activity in S. pombe.Citation19 During autophagy, Atg8-GFP is conjugated to the autophagosomal membrane and is broken down by proteolytic enzymes following fusion of the autophagosome and the vacuole, releasing free GFP. This GFP release can be detected by western blotting and, thus, used as a measure of autophagic activity.Citation22 Previous studies showed that nitrogen starvation induces autophagy in S. pombe, albeit much more slowly than in S. cerevisiae.Citation23 Hence, to test the induction of autophagy in S. pombe, Atg8-GFP-expressing cells lacking any auxotrophic markers were grown in either nitrogen-replete medium or nitrogen-free medium for 20 h. Importantly, western blot analysis revealed that GFP was released from the Atg8-GFP fusion protein when cells were starved of nitrogen but not during growth in nitrogen-replete medium, indicating that autophagy is induced by nitrogen starvation (, compare lanes 3 and 4). Furthermore, when cells were nitrogen-starved in media containing the vacuolar protease inhibitor PMSF, GFP release was greatly reduced (, compare lanes 4 and 6), confirming that Atg8-GFP is broken down following delivery to the vacuole. These data indicate that autophagy is responsible for the majority of Atg8-GFP breakdown during starvation and provide an assay for autophagy in fission yeast. Significantly, in agreement with previous work, Atg8-GFP hydrolysis was prevented in Δatg12 cells starved of nitrogen ().Citation19 Hence, if SpAtg10 functions as the E2 enzyme that facilitates Atg12 conjugation, SpAtg10 would also be essential for GFP release from the Atg8-GFP fusion protein during starvation. However, unexpectedly, GFP was released when Δatg10 cells were grown in nitrogen-free medium for 20 h (). Collectively, in contrast to Atg10 in S. cerevisiae,Citation11 these data suggest that SpAtg10 is not essential for autophagy in S. pombe.
Figure 2. A GFP release assay can be used to assess autophagy. (A and B) Cells lacking auxotrophic markers and expressing either untagged Atg8 (wt; SW576) or Atg8 tagged with GFP at the N terminus (Atg8-GFP; JT268) were incubated in minimal medium containing a nitrogen source (EMM) or minimal medium lacking a nitrogen source (EMM-N) in the presence or absence of 1 mM PMSF as indicated for 20 h at 30°C. Cell extracts were prepared and analyzed by western blotting using anti-GFP antibodies. The Atg8-GFP fusion protein and free GFP released during autophagy are indicated.
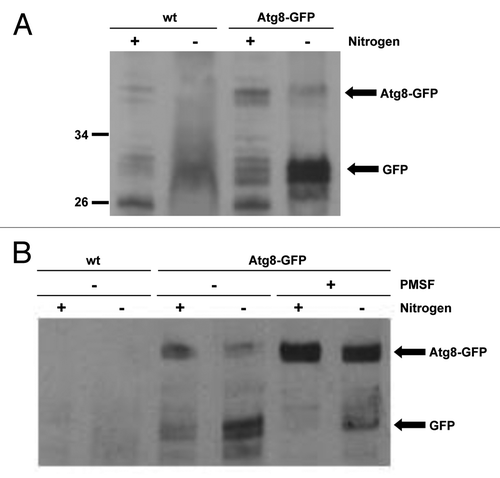
Figure 3. SpAtg10 is not essential for autophagy in S. pombe. (A) To investigate the potential role of SpAtg10 in autophagy, Atg8-GFP Δatg12 (MF43) and Atg8-GFP Δatg10 (MF31) cells lacking auxotrophic markers were incubated in EMM or EMM-N for 20 h at 30°C. Cell extracts were prepared and GFP release from the Atg8-GFP fusion protein was analyzed by western blotting using anti-GFP antibodies. The Atg8-GFP fusion protein and free GFP released during autophagy are indicated. (B) To determine whether SpAtg10 and/or Atg12 are required for the long-term survival of S. pombe cells during nitrogen starvation, wild-type (SW576), Δatg12 (MF13) and Δatg10 (MF15) cells were incubated in EMM-N medium at 30°C. Growth medium was removed and replaced with fresh medium every 3 d. Cells were removed from the EMM-N media and spotted onto YE5S plates after the indicated times. YE5S plates were incubated at 30°C for 3 d.
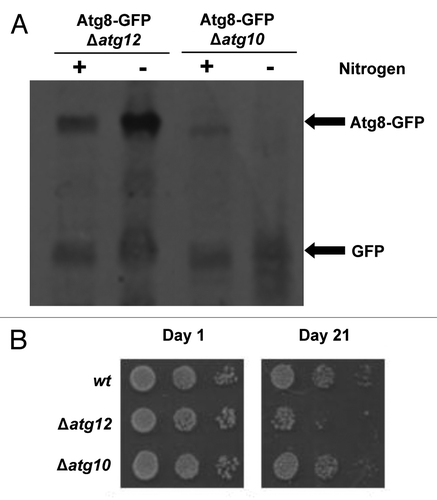
Autophagy is required for the long-term survival of fission yeast cells during nitrogen starvation, likely by providing an alternative nitrogen source via the degradation of proteins and organelles.Citation23 Although GFP release from Atg8-GFP is a good measure of autophagic activity, this assay does not provide an insight into how dysfunctional autophagy affects the long-term survival of S. pombe. Hence, we next assessed the potential role of SpAtg10 in the long-term survival of nitrogen-starved cells. After nitrogen starvation for up to 2 wk, Δatg12 and Δatg10 cells showed similar viability to wild-type cells (data not shown). However, following incubation for 3 wk, the viability of Δatg12 cells was significantly reduced compared with wild-type cells, demonstrating the importance of Atg12 for long-term survival during nitrogen starvation (). These results are in agreement with previous data showing that other atg mutants lose viability during long-term starvation.Citation23 However, in contrast to Δatg12 cells, the viability of Δatg10 cells was not reduced during starvation (). This is consistent with the results of the GFP release assay, which also suggested that SpAtg10 is not essential for autophagy. Although we cannot currently rule out a role for SpAtg10 in autophagy under certain conditions, our data strongly suggests that another E2 enzyme facilitates Atg12 conjugation during autophagy in S. pombe. One likely possible candidate for this missing E2 enzyme is Atg3. However, unfortunately, attempts to test this hypothesis were unsuccessful. Interestingly, although no thioester has been detected between the catalytic cysteine residue of Atg3 and Atg12, mammalian studies have demonstrated that Atg3 can facilitate Atg12 conjugation to Atg3Citation24 and is required for optimal Atg12-Atg5 conjugate formation.Citation25,Citation26 Furthermore, in contrast to S. cerevisiae, mammalian Atg10 is not essential for the conjugation of either Atg12 or Atg8.Citation27 Thus, these data raise the possibility that both Atg10 and Atg3 can act as E2 enzymes for Atg12 conjugation in certain eukaryotes. If this is the case, then it will be interesting to investigate whether Atg8 and Atg12 can compete for binding Atg3. However, taken together, these results support the conclusion that SpAtg10 is not essential for autophagy in S. pombe and, moreover, that SpAtg10 does not function as the E2 enzyme for Atg12.
SpAtg10 is essential for normal cell cycle progression
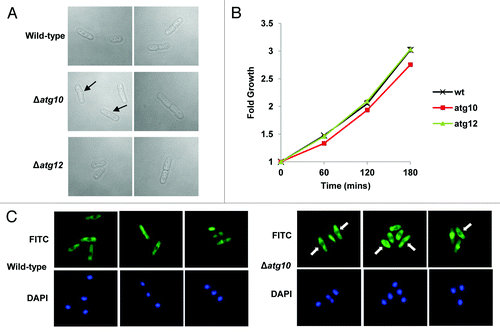
We next searched for alternative functions for SpAtg10. Interestingly, analysis of exponentially growing Δatg10 cells indicated that they appeared to be slightly longer than wild-type cells and, moreover, displayed other morphological defects (). In contrast, analysis of exponentially growing Δatg12 cells revealed similar morphologies to wild-type cells (). Indeed, consistent with these observations, analysis of wild-type, Δatg10 and Δatg12 cells using a coulter counter revealed mean cell volumes of 126.1 +/− 2.1 fl, 217.5 +/− 5.2 fl and 128.4 +/− 7.9 fl, respectively. In addition, similar analysis of wild-type, Δatg10 and Δatg12 cells demonstrated mean cell diameters of 5.96 +/− 0.04 μm, 7.17 +/− 0.05 μm and 5.98 +/− 0.09 μm, respectively. Finally, analysis of the growth of wild-type, Δatg10 and Δatg12 cells indicated that Δatg10 cells grow at a significantly slower rate than wild-type and Δatg12 cells (). Thus, collectively, these results indicate that SpAtg10, but not Atg12, is essential for normal cell cycle progression in S. pombe.
Figure 4. SpAtg10 but not Atg12 is essential for normal cell cycle progression. (A and B) Cells of mid-log-phase-growing cultures of wild-type (SW576), Δatg10 (MF15) and Δatg12 (MF13) strains, growing in liquid YE5S media at 30°C, were analyzed by (A) DIC microscopy and (B) a coulter counter. Growth curves were performed three times and error bars indicate the SEM. (C) Mid-log-phase-growing wild-type (CHP429) and Δatg10 (MF78) cells were fixed and spindle organization visualized by indirect immunofluorescence and nuclei visualized by DAPI staining. Representative images show cells of each strain at different stages of mitosis. In (A and C), the abnormal morphologies (small protrusions) of Δatg10 cells are indicated (arrows).
SpAtg10 is essential for responses to stress agents that perturb the cell cycle
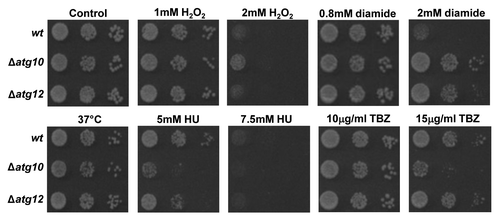
We next examined whether SpAtg10 and/or Atg12 is involved in cell responses to stress conditions that perturb the cell cycle. Similar to wild-type and Δatg12 cells, Δatg10 cells did not show temperature-sensitive growth at 37°C (). However, interestingly, Δatg10 cells displayed the greatest resistance to the oxidizing agents H2O2 and diamide but comparatively less resistance to the ribonucleotide reductase inhibitor HU, which blocks DNA replication (). In contrast to wild-type cells, Δatg10 cells also displayed increased sensitivity to the microtubule-depolymerizing agent TBZ, which prevents the formation of the mitotic spindle (). Interestingly, analysis of microtubule organization in mitotic cells revealed no major differences between wild-type and Δatg10 cells, revealing that the absence of SpAtg10 does not prevent the formation of microtubule fibers (). Recent studies have linked microtubules to the formation and trafficking of autophagosomes within the cell (for a review see ref. Citation28) and to trafficking autophagosomes during mitosis.Citation29 Hence, the increased TBZ sensitivity of Δatg10 cells raised the possibility that SpAtg10 functions as the E2 enzyme for Atg12 under certain conditions that are influenced by microtubule function. However, in marked contrast to Δatg10 cells, Δatg12 cells did not display any increased sensitivity to TBZ (), indicating that the increased sensitivity to TBZ demonstrated by Δatg10 cells is independent of Atg12 conjugation.
Figure 5. Loss of SpAtg10 affects the sensitivity of cells to drugs that perturb cell cycle progression. To investigate the potential role of SpAtg10 and the autophagy pathway in cell cycle processes, the sensitivity of wild-type (wt; SW576), Δatg10 (MF15) and Δatg12 (MF13) cells to different conditions known to affect cell cycle processes and checkpoints were examined. Ten-fold serial dilutions of mid-log-phase-growing cultures were spotted onto YE5S plates containing the indicated concentrations of H2O2, diamide, HU or TBZ. Plates were incubated at 30°C for 3 d. To investigate growth at 37°C cells were spotted onto a YE5S plate that was incubated at 37°C for 3 d.
Collectively, these data indicate that SpAtg10, the Atg10 homolog in S. pombe, is essential for normal cell cycle progression but not for autophagy. In addition, we can find no evidence that the autophagic pathway regulates cell cycle progression in a similar manner to SpAtg10. Homologs of Atg10 are found from yeast to human and, thus, our findings have important implications for the roles and regulation of Ubl conjugation pathways in other organisms. For example, Atg10 family proteins may have widespread roles independent of autophagy in the control of cell cycle progression. Furthermore, our work linking SpAtg10 to mitosis raises the possibility that Atg10 has dual roles in autophagy and mitosis in other organisms and may, in fact, have evolved to link these two fundamental processes.
Materials and Methods
Yeast strains and growth conditions
lists the S. pombe strains used in this study. Unless indicated otherwise, strains were grown at 30°C in either YE5S (rich) or EMM (minimal) media.Citation30 To induce autophagy, strains were grown in EMM lacking a nitrogen source (EMM-N) for the indicated times.Citation23 Mean cell volumes and mean cell diameters were calculated using a coulter counter (Schärfe System) and are presented +/− 3 standard deviations.
Table 2. The S. pombe strains used in this study
Strain constructions
To generate Δatg10 strains, a deletion cassette, where the atg10+ gene was replaced by KanMX, was first obtained by PCR using the oligonucleotide primers Atg10delf (5′-GTTGGATAACTTTGTAAACTCC-3′) and Atg10delr (5′-GTGGAACAGTTGAATGATATG-3′) and genomic DNA from BG_0880 as a template. The deletion cassette was then introduced into SW576, JT268 and CHP429, to generate MF15, MF31 and MF78, respectively. To generate Δatg12 strains, a deletion cassette, where the atg12+ gene was replaced by KanMX, was first obtained by PCR using the oligonucleotide primers Atg12delf (5′-GTGACAGGAAAGTTGATGC-3′) and Atg12delr (5′-CTGTCTAAGGCTGACGCTGC-3′) and genomic DNA from BG_0465 as a template. The deletion cassette was then introduced into SW576 and JT268 to generate MF13 and MF43, respectively. All strain constructions were confirmed by PCR.
Autophagy induction and analyses
To induce autophagy, strains were grown to mid-log phase in EMM, washed three times in EMM-N and finally resuspended in a volume of EMM-N equivalent to the original volume of the culture. To assess autophagy, cells were starved of nitrogen for 20 h, then extracts were prepared as described and analyzed by western blotting as described below. To investigate long-term survival during nitrogen starvation, cells were incubated in EMM-N for the indicated times at 30°C. During starvation, the growth medium was replaced with fresh EMM-N every 3 d. To determine viability at the indicated time points, samples of cultures were diluted, plated onto solid YE5S medium and incubated at 30°C for 3 d.
Sensitivity tests
~Five μl of 10-fold serial dilutions of mid-log-phase-growing cultures were spotted onto solid YE5S medium containing the indicated concentrations of H2O2, diamide, hydroxyurea (HU) and thiabendazole (TBZ). Plates were incubated at 30°C for 3 d.
Protein extracts and western blot analyses
Cultures were grown and extracts prepared using lysis buffer (50 mM TRIS-HCl pH 7.5, 150 mM NaCl, 0.5% Nonidet P-40, 10 mM imidazole, 0.1% leupeptin, 0.1% pepstatin A, 1% aprotinin, 1% PMSF, 0.2% Na3VO4, 5% NaF).Citation31 Protein samples were then analyzed by SDS-PAGE and transferred onto a nitrocellulose transfer membrane (Protran®). To investigate autophagic activity, the levels of Atg8-GFP and free GFP were examined using a 1:2,000 dilution of mouse anti-GFP antibody (Invitrogen) and a 1:2,000 dilution of anti-mouse HRP-conjugated secondary antibody (Sigma-Aldrich). Proteins were visualized using the ECLTM detection system (Amersham Pharmacia Biotech).
Fluorescence microscopy
To analyze microtubules in mitotic cells, cells were fixed and prepared for microscopy as described previously.Citation31 To visualize microtubules, a 1:1,000 dilution of anti-TAT1 antibody (Cancer Research UK) and a 1:200 dilution of secondary antibody [Alexa Fluor® FITC-conjugated anti-mouse antibody (Invitrogen)] were used for indirect immunofluorescence. Cells were mounted onto poly-L-lysine-coated microscope slides with Vectashield mounting medium (containing DAPI-Vector Laboratories). FITC/DAPI-treated cells were excited with the appropriate wavelengths of light using a Zeiss Axiovision microscope and visualized with an Axiovision imaging system.
| Abbreviations: | ||
| DAPI | = | 4’-6-diamidino-2-phenylindole |
| DIC | = | differential interference contrast |
| FITC | = | fluorescein isothiocyanate |
| GFP | = | green fluorescent protein |
| HRP | = | horse-radish peroxidase |
| HU | = | hydroxyurea |
| PCR | = | polymerase chain reaction |
| PMSF | = | phenylmethylsulfonyl fluoride |
| TBZ | = | thiabendazole |
| Ub/Ubl | = | ubiquitin/ubiquitin-like protein |
Acknowledgments
We thank Viktor Korolchuk, Ellen Rumsby, Elizabeth Veal, Masayuki Yamamoto and Bioneer for kindly providing either strains or comments on the manuscript, and Michelle Wray for technical assistance. The work was funded by the Biotechnology and Biological Sciences Research Council (M.F. PhD studentship).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Hochstrasser M. Evolution and function of ubiquitin-like protein-conjugation systems. Nat Cell Biol 2000; 2:E153 - 7; http://dx.doi.org/10.1038/35019643; PMID: 10934491
- Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol 2006; 22:159 - 80; http://dx.doi.org/10.1146/annurev.cellbio.22.010605.093503; PMID: 16753028
- Reggiori F, Klionsky DJ. Autophagy in the eukaryotic cell. Eukaryot Cell 2002; 1:11 - 21; http://dx.doi.org/10.1128/EC.01.1.11-21.2002; PMID: 12455967
- Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 1993; 333:169 - 74; http://dx.doi.org/10.1016/0014-5793(93)80398-E; PMID: 8224160
- Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci 2005; 118:7 - 18; http://dx.doi.org/10.1242/jcs.01620; PMID: 15615779
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol 2009; 10:458 - 67; http://dx.doi.org/10.1038/nrm2708; PMID: 19491929
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol 2007; 9:1102 - 9; http://dx.doi.org/10.1038/ncb1007-1102; PMID: 17909521
- Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ 2005; 12:Suppl 2 1542 - 52; http://dx.doi.org/10.1038/sj.cdd.4401765; PMID: 16247502
- Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep 2008; 9:859 - 64; http://dx.doi.org/10.1038/embor.2008.163; PMID: 18704115
- Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, et al. A protein conjugation system essential for autophagy. Nature 1998; 395:395 - 8; http://dx.doi.org/10.1038/26506; PMID: 9759731
- Shintani T, Mizushima N, Ogawa Y, Matsuura A, Noda T, Ohsumi Y. Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J 1999; 18:5234 - 41; http://dx.doi.org/10.1093/emboj/18.19.5234; PMID: 10508157
- Tanida I, Mizushima N, Kiyooka M, Ohsumi M, Ueno T, Ohsumi Y, et al. Apg7p/Cvt2p: A novel protein-activating enzyme essential for autophagy. Mol Biol Cell 1999; 10:1367 - 79; PMID: 10233150
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, et al. A ubiquitin-like system mediates protein lipidation. Nature 2000; 408:488 - 92; http://dx.doi.org/10.1038/35044114; PMID: 11100732
- Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, et al. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol 1999; 147:435 - 46; http://dx.doi.org/10.1083/jcb.147.2.435; PMID: 10525546
- Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, et al. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol 2000; 151:263 - 76; http://dx.doi.org/10.1083/jcb.151.2.263; PMID: 11038174
- Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem 2007; 282:37298 - 302; http://dx.doi.org/10.1074/jbc.C700195200; PMID: 17986448
- King RW, Deshaies RJ, Peters JM, Kirschner MW. How proteolysis drives the cell cycle. Science 1996; 274:1652 - 9; http://dx.doi.org/10.1126/science.274.5293.1652; PMID: 8939846
- Mizushima N, Yoshimori T, Ohsumi Y. Mouse Apg10 as an Apg12-conjugating enzyme: analysis by the conjugation-mediated yeast two-hybrid method. FEBS Lett 2002; 532:450 - 4; http://dx.doi.org/10.1016/S0014-5793(02)03739-0; PMID: 12482611
- Mukaiyama H, Kajiwara S, Hosomi A, Giga-Hama Y, Tanaka N, Nakamura T, et al. Autophagy-deficient Schizosaccharomyces pombe mutants undergo partial sporulation during nitrogen starvation. Microbiology 2009; 155:3816 - 26; http://dx.doi.org/10.1099/mic.0.034389-0; PMID: 19778961
- Mukaiyama H, Nakase M, Nakamura T, Kakinuma Y, Takegawa K. Autophagy in the fission yeast Schizosaccharomyces pombe. FEBS Lett 2010; 584:1327 - 34; http://dx.doi.org/10.1016/j.febslet.2009.12.037; PMID: 20036658
- Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, Sekido S, et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol 2006; 24:841 - 7; http://dx.doi.org/10.1038/nbt1222; PMID: 16823372
- Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy 2007; 3:181 - 206; http://dx.doi.org/10.4161/auto.3678; PMID: 17224625
- Kohda TA, Tanaka K, Konomi M, Sato M, Osumi M, Yamamoto M. Fission yeast autophagy induced by nitrogen starvation generates a nitrogen source that drives adaptation processes. Genes Cells 2007; 12:155 - 70; http://dx.doi.org/10.1111/j.1365-2443.2007.01041.x; PMID: 17295836
- Radoshevich L, Murrow L, Chen N, Fernandez E, Roy S, Fung C, et al. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell 2010; 142:590 - 600; http://dx.doi.org/10.1016/j.cell.2010.07.018; PMID: 20723759
- Tanida I, Tanida-Miyake E, Komatsu M, Ueno T, Kominami E. Human Apg3p/Aut1p homologue is an authentic E2 enzyme for multiple substrates, GATE-16, GABARAP, and MAP-LC3, and facilitates the conjugation of hApg12p to hApg5p. J Biol Chem 2002; 277:13739 - 44; http://dx.doi.org/10.1074/jbc.M200385200; PMID: 11825910
- Sou YS, Waguri S, Iwata JI, Ueno T, Fujimura T, Hara T, et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell 2008; 19:4762 - 75; http://dx.doi.org/10.1091/mbc.E08-03-0309; PMID: 18768753
- Nemoto T, Tanida I, Tanida-Miyake E, Minematsu-Ikeguchi N, Yokota M, Ohsumi M, et al. The mouse APG10 homologue, an E2-like enzyme for Apg12p conjugation, facilitates MAP-LC3 modification. J Biol Chem 2003; 278:39517 - 26; http://dx.doi.org/10.1074/jbc.M300550200; PMID: 12890687
- Monastyrska I, Rieter E, Klionsky DJ, Reggiori F. Multiple roles of the cytoskeleton in autophagy. Biol Rev Camb Philos Soc 2009; 84:431 - 48; http://dx.doi.org/10.1111/j.1469-185X.2009.00082.x; PMID: 19659885
- Xie R, Nguyen S, McKeehan WL, Liu L. Acetylated microtubules are required for fusion of autophagosomes with lysosomes. BMC Cell Biol 2010; 11:89; http://dx.doi.org/10.1186/1471-2121-11-89; PMID: 21092184
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 1991; 194:795 - 823; http://dx.doi.org/10.1016/0076-6879(91)94059-L; PMID: 2005825
- Bulmer R, Pic-Taylor A, Whitehall SK, Martin KA, Millar JB, Quinn J, et al. The forkhead transcription factor Fkh2 regulates the cell division cycle of Schizosaccharomyces pombe.. Cell 2004; 3:944 - 54; http://dx.doi.org/10.1128/EC.3.4.944-954.2004; PMID: 15302827