 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
The neonatal Fc receptor (FcRn) protects immunoglobulin G (IgG) from degradation and increases the serum half-life of IgG, thereby contributing to a higher concentration of IgG in the serum. Because altered FcRn binding may result in a reduced or prolonged half-life of IgG molecules, it is advisable to characterize Fc receptor binding of therapeutic antibody lead candidates prior to the start of pre-clinical and clinical studies.
In this study, we characterized the interactions between FcRn of different species (human, cynomolgus monkey, mouse and rat) and nine IgG molecules from different species and isotypes with common variable heavy (VH) and variable light chain (VL) domains. Binding was analyzed at acidic and neutral pH using surface plasmon resonance (SPR) and biolayer interferometry (BLI).
Furthermore, we transferred the well-accepted, but low throughput SPR-based method for FcRn binding characterization to the BLI-based Octet platform to enable a higher sample throughput allowing the characterization of FcRn binding already during early drug discovery phase. We showed that the BLI-based approach is fit-for-purpose and capable of discriminating between IgG molecules with significant differences in FcRn binding affinities.
Using this high-throughput approach we investigated FcRn binding of 36 IgG molecules that represented all VH/VL region combinations available in the fully human, recombinant antibody library Ylanthia®. Our results clearly showed normal FcRn binding profiles for all samples. Hence, the variations among the framework parts, complementarity-determining region (CDR) 1 and CDR2 of the fragment antigen binding (Fab) domain did not significantly change FcRn binding.
Introduction
In their application as protein therapeutics, antibodies have been proven to offer successful treatment options for a large variety of human diseases.Citation1,Citation2 The pharmacokinetics (PK) of antibodies is influenced by several factors, e.g., charge and glycosylation of the antibody, target affinity, expression and biology, injection route, neonatal Fc receptor (FcRn) binding.Citation3 As a receptor of immunoglobulin G (IgG) molecules, the FcRn is responsible for the transfer of IgGs from a mother to the fetus.Citation4,Citation5 In addition, FcRn protects IgGs from degradation and increases the serum half-life, and in consequence also the serum concentration, of IgGs.Citation6 This was also shown for albumin whose half-life is extended by FcRn activity as well.Citation7,Citation8 Altered FcRn binding may result in a reduced or prolonged half-life of IgG molecules.Citation9,Citation10
FcRn is expressed by endothelial cells, which internalize serum components including soluble IgGs from the bloodstream by pinocytosis. IgG binding to FcRn is pH-dependent;Citation11 the acidic pH (pH 6.0) inside the endosomal compartment allows the IgGs to bind to FcRn. After recycling back to the cell surface, the IgG dissociates from FcRn at physiological pH (~pH 7.2), is released back into the blood circulation and thereby protected from lysosomal degradation.Citation4
FcRn is a major histocompatibility complex class I-like heterodimer composed of the soluble light chain β2-microglobulin (β2m) and a membrane-bound heavy chain.Citation12 Crystal structure analysis revealed that rat FcRn (rFcRn)Citation12,Citation13 and human FcRn (hFcRn)Citation14 bind to the CH2-CH3 hinge region of both heavy chains of the Fc homodimer of an IgG, resulting in a 2:1 stoichiometry.Citation12,Citation13 The interaction between FcRn and Fc is mainly stabilized by salt bridges between anionic FcRn residues and histidine residues of the IgG, which are protonated at acidic pH.Citation15,Citation16 A detailed review has been published by Roopenian and Akilesh.Citation4
When performing in vivo experiments, the cross-species binding between FcRn and IgG must be considered.Citation10,Citation17 The hFcRn can bind a limited set of IgG molecules from primates and rabbits, but not rodent IgGs.Citation18,Citation19 The mouse FcRn (mFcRn) can bind IgGs from various species including human.Citation20 Additionally, pH-dependency of binding, as well as absolute affinities, differ between mFcRn and hFcRn.Citation19,Citation21 To overcome this limitation of animal models in terms of pharmacokinetics (PK) comparability, a transgenic mouse model is available, in which hFcRn is expressed instead of the mouse ortholog.Citation22 This model can be used to test human IgGsCitation9,Citation23 with the limitation that the overall mouse IgG level is strongly decreased.Citation24 Using this mouse model, in some cases a correlation between the PK of IgGs in primates, humans, and mice was observed.Citation22,Citation25
Several studies were performed to investigate the in vitro affinity of various IgG formats from different species to FcRn molecules. FcRn molecules are not commercially available, but their production in various cells, such as in Chinese hamster ovary,Citation14,Citation26 human cell lines such as HEK293Citation27 and PEAK cells (human embryo kidney monolayer epithelial cells)Citation19 or E. coliCitation28 has been recently published. However, the published FcRn binding results differ significantly. Some major causes contributing to these differences are probably the different FcRn materials, assay formats and evaluation models used.Citation25 Beside ELISA measurementsCitation29,Citation30 or cell-based assaysCitation31,Citation32 real-time measurements using surface plasmon resonance (SPR)Citation19,Citation23,Citation30,Citation33,Citation34 are often performed to characterize in vitro binding. Published SPR results have been generated using different experimental setups and evaluation strategies, and consequently also the reported affinities vary significantly.Citation25 For instance, for binding of an IgG1 to hFcRn, apparent KD values from 6 to 2500 nM have been reported.Citation9,Citation14,Citation19,Citation23,Citation25,Citation27,Citation35-Citation37 This makes a literature-based comparison of reported affinities very challenging and hampers interpretation of results.Citation23,Citation38
Deng et al.Citation39 showed that the binding pattern for one humanized mouse IgG to cynomolgus monkey FcRn (cyFcRn) and hFcRn was comparable. The affinity of cyFcRn was reported to be ~2-fold higher than that of hFcRn. Equal observations were made by Dall’AcquaCitation36 and Datta-Mannan.Citation37 However, to our knowledge, a detailed binding study of different IgG isotypes and species to cyFcRn is not yet available.
Antibodies with extended half-life would enable more appropriate clinical dosing, a major benefit for the patients. Although the average half-life of a human IgG is 21 d,Citation40 IgGs with wild type Fc sequences can have a considerably lower half-life of down to 8 d.Citation41 Therefore, it would be useful to adequately estimate the in vivo half-life of an antibody, based on the results from in vitro experiments. Accordingly, several publications compared in vitro FcRn binding to the in vivo half-life of monoclonal antibodies, but the results were mixed, i.e., a correlation between FcRn affinity and half-life was shown by several workgroups,Citation9,Citation31,Citation39,Citation42 whereas others could not see a direct correlation.Citation25,Citation36,Citation43 A recent publication of Wang et al.Citation23 compared the in vitro FcRn binding of different IgGs containing wild type human Fc sequences with their in vivo PK behavior. For the affinity (i.e., association and dissociation) determined at acidic pH, no correlation was observed. However, when the dissociation was analyzed at neutral pH and used for comparison, a correlation between sufficient release at neutral pH and extended half-life was observed. As the Fc parts of the IgGs were identical, the authors suggested that the antigen binding fragment (Fab) may also affect the FcRn interaction.Citation23 Igawa and coworkers showed that engineering of the variable region may also enhance the half-life in an FcRn-independent way.Citation27,Citation44,Citation45
Several groups applied amino acid point mutations within the FcRn binding site of the IgG, resulting in an increased FcRn affinity at pH 6.0 as reviewed by Kuo et al.Citation10 In several cases, these IgGs had an improved half-life in animal studies.Citation9,Citation25,Citation31,Citation39 Nevertheless, point mutations causing a general increase in affinity at both, pH 6.0 and pH 7.2 could also result in decreased half-life.Citation35,Citation37,Citation39 This observation is in good agreement with the results of Wang et al.,Citation23 who showed that the measured dissociation at neutral pH could be correlated with the in vivo PK, i.e., decreased dissociation correlated with shortened half-life.
The analysis of the in vitro binding of IgG to FcRn will be beneficial for early exclusion of lead candidates with unfavorable PK behavior.Citation10,Citation23 To generate a comprehensive data set on FcRn binding, we performed a detailed study using FcRn from different relevant species (human, cynomolgus monkey, mouse, rat), and IgG samples with different isotypes from these species. All the IgG samples shared a common variable region, to exclude the influence of this region on the determined affinities. Differences in FcRn binding of the different IgG isotypes and species could clearly be observed and quantified.
To further investigate the importance of determinants outside of the constant regions, i.e., of variable domain framework and complementarity-determining regions (CDR), we performed additional FcRn binding experiments. To this end, we investigated FcRn interactions of a sample set consisting of 36 fully human IgG1 antibodies. All the IgG samples only differed in their variable region VH/VL combinations (from N-terminus through framework 3, ). These molecules were selected as scaffolds for the fully human antibody library Ylanthia®, which was designed and proven to generate antibodies with favorable biophysical properties from the outset, without the need for further optimization.Citation46 For this IgG1 sample set, we did not observe significant differences in binding affinities to human, cynomolgus monkey, rat and mouse FcRn molecules.
Figure 1. Schematic representation of IgG molecule classes analyzed in this study. Shown is the overall structure of different anti-lysozyme IgG molecules (A) and Ylanthia® IgG1 scaffolds (B) characterized for binding to human, cynomolgus monkey, rat and mouse FcRn. Parts of the molecules which were identical in the respective sample set are shown in gray color, whereas varied IgG parts are shown in blue (A) and green (B) color. For the nine anti-lysozyme molecules (MOR03207, A), the variable regions (VH/VL) were identical, whereas constant regions (CL, CH1, CH2, CH3) belongs to IgG variants and species variants as indicated below the figure. For the 36 Ylanthia® IgG1 scaffolds (B), the constant regions of heavy and light chain were identical, as well as the CDR-H3 and framework 4 of heavy chain and kappa CDR-L3 and lambda CDR-L3 and the respective framework 4 (for details, refer to Ewert et al.Citation60). Other regions in the VH and VL domains represented respective sequences from 12 different VH, 9 Vκ, and 6 Vλ genes.
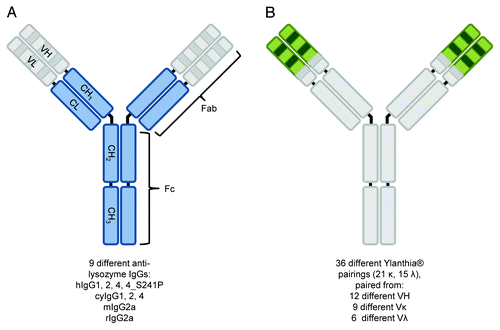
In addition, we found it desirable to establish a high throughput method for processing this large sample set. In this report, we describe the successful transfer of a rather low throughput method for determining FcRn affinities using SPR to a biolayer interferometry (BLI) platform, which resulted in a generally applicable, robust, high throughput method for profiling of FcRn binding.
Results
Production and characterization of FcRn molecules
To perform the intended FcRn binding characterization experiments with a comprehensive sample set of high quality, FcRn molecules (human, cynomolgus monkey, rat, and mouse sequences) were cloned, expressed and purified using comparable protocols. To enable site-specific biotinylation and a one-step purification of the heterodimeric FcRn molecules, an avi-His-tag was fused to the C-terminal part of the membrane-bound heavy chain (α-chain), which showed a lower expression level in comparison to the β2-microglobulin (β2m). Therefore, the β2m was present in surplus in the cell culture supernatant and IMAC-purification was used to separate the heterodimer from free β2m. Yields of the hFcRn and cyFcRn molecules were ~28 mg/L (mg purified FcRn per liter of culture volume) and 18 mg/L, while the yields of the mFcRn and rFcRn were ~5 mg/L and 2.5 mg/L, respectively.
All four FcRn molecules showed comparable high qualities as assessed by SDS-PAGE under reducing and non-reducing conditions, analytical HP-SEC-MALS and dynamic light scattering (DLS, ). The α-chain of the rodent FcRn appeared larger than that of the primate FcRn in SDS-PAGE (). The observed shift in apparent molecular weight from ~38 to 50 kDa is probably due to the higher number of N-linked glycosylation sites of the rodent FcRn molecules. The β2m of all four FcRn molecules were visible at ~12 kDa in SDS-PAGE.
Figure 2. Characterization of human, cynomolgus monkey, mouse and rat FcRn by SDS-PAGE (A), analytical HP-SEC-MALS (B) and DLS (C). The characterization of all four FcRn molecules showed high purity, homogeneity and correct molecular weights for all FcRn preparations (see Results for details). red., reducing conditions; non-red., non-reducing conditions.
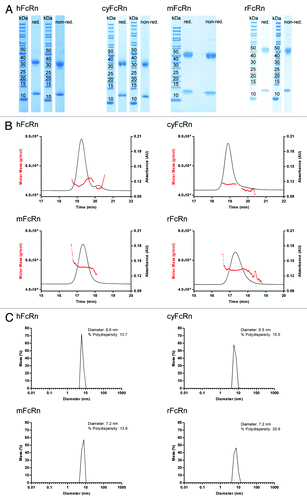
HP-SEC-MALS analysis revealed a pure preparation with only one species with a molecular mass of 47 kDa as determined by multi-angle light scattering for the hFcRn and cyFcRn, 55 kDa for the mFcRn and 54 kDa for the rFcRn (), respectively, which is in agreement with the masses calculated from the primary sequences. This confirmed that the samples contained only FcRn heterodimers because no larger or smaller species were detected.
All purified FcRn samples were essentially monodisperse and did not contain higher-order aggregates (which would not be visible in HP-SEC) as determined by DLS ().
Comparative binding of different IgG molecules to human FcRn as determined by SPR
Different SPR assay setups have been described in the literature, where either the receptor or IgG was used as immobilized ligand on the chip surface. When using FcRn as ligand, covalent immobilization onto the sensor chipCitation42 or binding of biotinylated FcRn on a streptavidin surfaceCitation19 have been described, followed by injection of IgG as the analyte. Similar to the setup described by Magistrelli and coworkers,Citation19 we used biotinylated FcRn bound as ligand onto a streptavidin sensor chip and IgG samples were used as analytes.
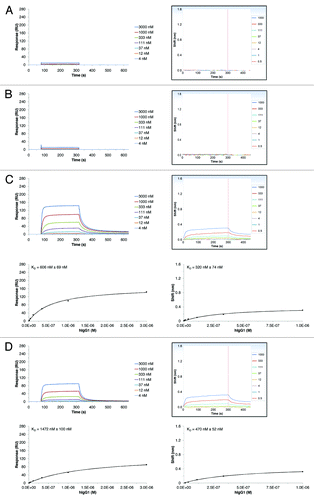
The SPR assay was used to examine the in vitro binding of different IgG isotypes and species on hFcRn. The analysis was performed with variants of an anti-lysozyme antibody (MOR03207). While all investigated MOR03207 antibody variants share identical variable (V) region sequences, their constant regions differ depending on the IgG isotype or species. For this study, four human IgG variants (hIgG1, hIgG2, hIgG4, hIgG4_S241P), three cynomolgus monkey variants (cyIgG1, cyIgG2, cyIgG4), one mouse (mIgG2a) and one rat (rIgG2a) variant of MOR03207 were used. The human IgG3 isotype was regarded to be of minor therapeutic relevance and therefore was omitted from the comparative analysis. A set of typical sensorgrams of IgG variants interacting with hFcRn at pH 6.0 or pH 7.2 is shown in (examples shown: cyIgG1, cyIgG2, or cyIgG4; see Supplemental Material for complete compendium of sensorgrams of all analyzed interactions). Steady-state fit analysis was performed as described in Materials and Methods. The resulting steady-state fits were plotted () and the apparent equilibrium dissociation constants (in the following named KD) were calculated (). In our experiments, all tested hIgG and cyIgG isotypes bound to hFcRn at pH 6.0. In contrast, mouse and rat IgG2a did not bind to hFcRn at pH 6.0, which is consistent with a publication of Ober et al.Citation18 As expected, all tested IgG molecules showed no or negligible binding to hFcRn at pH 7.2 even at the highest concentration analyzed (3000 nM). In summary, our results clearly indicate that the IgG isotype has a negligible influence on hFcRn binding, which is in line with previous findings.Citation23
Figure 3. SPR results of the interaction between different cyIgG molecules and hFcRn. SPR sensorgrams for cyIgG1 (A), cyIgG2 (B) and cyIgG4 (C) were recorded at pH 6.0 (left panels) and pH 7.2 (right panels). The IgG-FcRn KD values (given errors represent 95% confidence intervals of the fits) were determined using a steady-state model, corresponding fits at pH 6.0 are presented in the lower panels. Response scale of ordinates was kept constant for all sensorgrams and steady-state figures throughout the manuscript to enable a direct comparison.
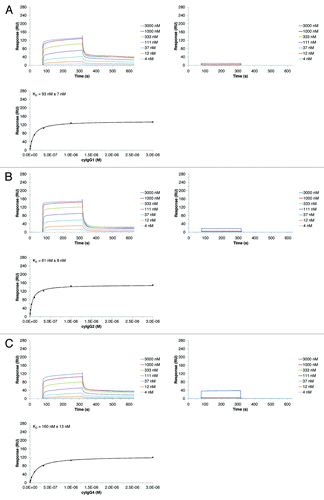
Table 1. Summary of determined equilibrium dissociation constants (KD) of interaction of IgG with FcRn molecules
Comparative binding of IgG to human, cynomolgus monkey, mouse and rat FcRn as determined by SPR
To assess the cross-specificity of the various IgG isotype, we examined the binding of IgG samples to FcRn orthologs from human, cynomolgus monkey, mouse and rat by SPR (, left panels and Supplemental Materials). The results shown in , as well as previously published data, demonstrate that hFcRn is highly species-selective, binding only the primate IgGs and not rodent IgGs.Citation18 The same species-selective binding behavior was observed for the interaction between the tested IgGs and cyFcRn. CyFcRn and hFcRn also revealed similar results at pH 7.2, i.e., no or negligible binding (, left panel). None of the investigated IgG isotypes bound to hFcRn or cyFcRn at neutral pH. Representative sensorgrams in (left panel) show the negligible binding of hIgG1 to hFcRn and cyFcRn at pH 7.2.
Figure 4. Comparison of SPR (left panels) and BLI (right panels) assay results of the interaction between hIgG1 and different FcRn species. The binding kinetics for hIgG1 to human (A), cynomolgus monkey (B), mouse (C) and rat (D) FcRn were recorded on both platforms at pH 6.0. The IgG-FcRn KD values were determined using a steady-state model. Given errors represent 95% confidence intervals of the fits.
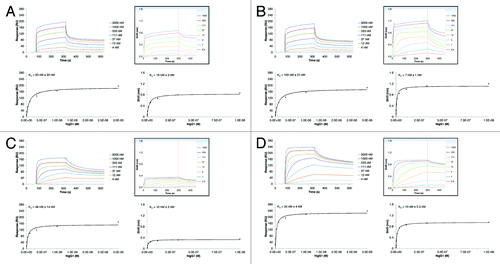
Figure 5. Comparison of SPR (left panels) and BLI (right panels) assay results of the interaction between hIgG1 and different FcRn species. The binding kinetics for hIgG1 to human (A), cynomolgus monkey (B), mouse (C) and rat (D) FcRn were recorded on both platforms at pH 7.2. For human and cynomolgus monkey negligible binding was detected, for mouse and rat FcRn, the IgG-FcRn KD values were determined using a steady-state model. The given errors represent 95% confidence intervals of the fits.
All tested IgG isotypes interacted well with mFcRn and rFcRn at pH 6.0. Compared with the interactions observed with primate FcRn molecules, the resulting binding curves were less heterogeneous and showed markedly slower dissociation. Consequently the evaluation of hIgG and cyIgG isotypes resulted in better affinities (i.e., lower KD) to mFcRn and rFcRn compared with hFcRn and cyFcRn, which is in line with previously published data.Citation18,Citation39 In contrast to the hFcRn and cyFcRn, which did not bind rodent IgGs at pH 6.0 and pH 7.2, mFcRn and rFcRn interacted with the tested human and cynomolgus IgG isotypes at pH 7.2. Nevertheless, the resulting steady-state KD values were significantly higher compared with pH 6.0. In SPR experiments at neutral pH, no binding was detected for mIgG2a or rIgG2a to any of the investigated receptors (human, cynomolgus monkey, mouse, or rat FcRn).
Development of an FcRn screening platform using BLI and comparison with SPR results
To enable a flexible screening platform with increased throughput, we adapted the IgG/FcRn interaction assay to the PALL/ForteBio Octet QK384 platform using essentially the same assay set up and steady-state fit model as for the SPR platform. The Octet QK384 system comprises 16 parallel, independent sensors to measure, e.g., protein-protein interactions in real-time, using BLI as the detection principle. BLI is based on interferometry, i.e., it is a label-free method to measure interactions of soluble analytes with a sensor surface, onto which the corresponding interaction partner is immobilized. In contrast to SPR, no microfluidics are used in Octet instruments, but the biosensors move to the samples held in an open shaking 96- or 384-well microplate.Citation47
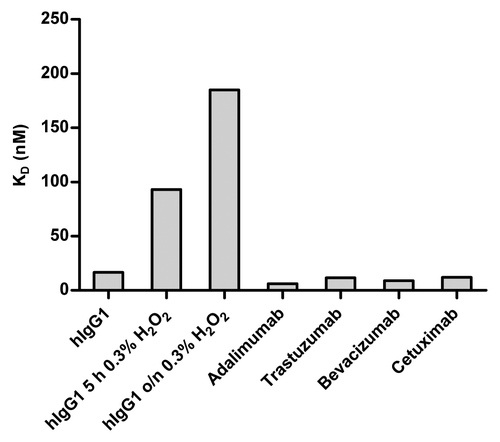
In a first step of testing the performance of the BLI-based method, we analyzed the interaction of different IgG molecules with hFcRn at pH 6.0. To investigate the assay’s ability to discriminate between different affinities, several marketed therapeutic IgG molecules (adalimumab, trastuzumab, bevacizumab, and cetuximab) and hIgG1 (MOR03207, anti-lysozyme antibody) were examined. Since it has been reported that oxidation of hIgG1 (i.e., of exposed methionines at position 252 and 428) significantly decreases the binding affinity to hFcRn at pH 6.0,Citation48 we included such a control in the assay. Human IgG1 (MOR03207) was incubated with 0.3% H2O2 at room temperature for 5 h or overnight, respectively, according to Liu et al.Citation49 As expected from published results,Citation23 measured affinities for marketed therapeutic IgG molecules were largely comparable (steady-state affinity values in the range of 6 to 17 nM) (). In good agreement with results from Wang et al.,Citation48 we observed a 5-fold reduced binding affinity for the oxidized IgG1 sample that was treated with H2O2 for 5 h compared with the untreated version. The effect was even more pronounced when the sample was exposed to H2O2 overnight (~11-fold reduction). These results show that the developed BLI-based method is fit-for-purpose, i.e., it is capable of discriminating between different FcRn binding affinities.
Figure 6. Determined equilibrium dissociation constants (KD) of interaction of hIgG1, oxidized hIgG1 and different marketed therapeutic antibodies with hFcRn. Binding of IgG to hFcRn at pH 6.0 was determined by BLI. Biotinylated FcRn was immobilized on the streptavidin sensor tips and IgG molecules were used as analytes. Data were recorded on a PALL/ForteBio Octet QK384 instrument (see Materials and Methods for details). KD values of MOR03207 hIgG1 and marketed therapeutic antibodies were comparable (6 to 17 nM). As a control, to assess the capability of the BLI-based method to discriminate between different affinities, hIgG1 (MOR03207) was oxidized by exposure to 0.3% (v/v) H2O2 for 5 h and overnight incubation. As expected, the affinities of the oxidized IgG samples were strongly reduced (KD ~11-fold increased after overnight incubation).
To further characterize the BLI-based method, different species, isotypes and variants of anti-lysozyme antibody MOR03207 were used as a representative panel of IgG molecules in order to compare the results directly with the results generated using the SPR-based approach. The equilibrium dissociation constants of hIgG1, hIgG4, hIgG4_S241P, mIgG2a and rIgG2a were determined on human, cynomolgus monkey, mouse and rat FcRn at pH 6.0. The results are shown in . In general, the results obtained by both methods were qualitatively in good agreement. However, the KD values determined by SPR were up to 7-fold higher compared with the KD values determined by BLI, except for IgG binding to cynomolgus monkey FcRn, where SPR KD values were approximately up to 19-fold higher than the KD values obtained by BLI. But the relative differences between KD results of the different samples were comparable between both methods. The BLI results showed no major affinity differences between the analyzed FcRn species, i.e., KD values varied in a range from 6 to 27 nM, except for mIgG2a and rIgG2a, which did not bind to human or cynomolgus FcRn. No obvious trend in KD for FcRn of different species was visible. In contrast, the SPR results showed slightly lower KD values for mFcRn and rFcRn in comparison to hFcRn and cyFcRn (2- to 3-fold). Similar to the BLI results, mIgG2a and rIgG2a did not bind to human and cynomolgus FcRn.
Figure 7. Comparison of determined equilibrium dissociation constants (KD) of the interaction of different IgG molecules with FcRn from different species at pH 6.0 and pH 7.2. Binding of different MOR03207 IgG molecules to human (h), cynomolgus monkey (cy), mouse (m) and rat (r) FcRn at pH 6.0 (A) and pH 7.2 (B) was determined by SPR (white bars) and BLI (black bars; see Materials and Methods for details). Qualitative binding results were in good agreement, however, absolute KD values differed by a factor of up to ~7 between different methods at both pH values analyzed except for binding to cyFcRn. Here, KD values by SPR were up to ~19-fold increased compared with BLI results at pH 6.0. For details see Results and Discussion. NB, no or negligible binding observed.
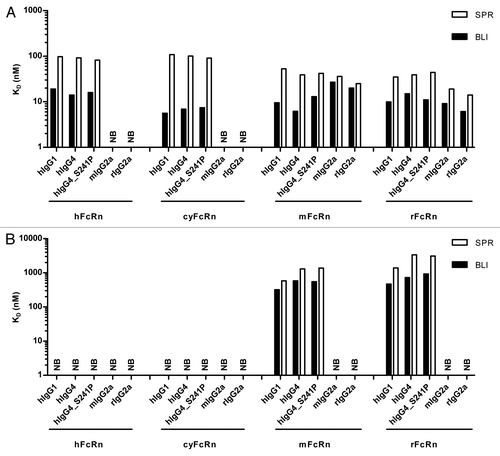
In addition to binding at pH 6.0, it is crucial to analyze the IgG-FcRn interactions at neutral pH, which appear to be independent of pH 6.0 binding affinities.Citation23 As shown in , binding of hIgG1 to mouse and rat FcRn could be demonstrated with SPR as well as with BLI, whereas no or negligible binding was detected to human and cynomolgus FcRn at pH 7.2. Similar results were obtained for hIgG4 and hIgG4_S241P. Consistent with the observations at pH 6.0, the KD values determined by SPR were up to 5-fold higher compared with the KD values determined by BLI. In both methods, human IgGs showed binding to rodent FcRn, although the affinities were consistently lower at neutral pH than at pH 6.0. In both, SPR and BLI analysis, mouse and rat IgG2a showed no or negligible binding to any of the tested FcRn species at pH 7.2.
Analysis of FcRn binding of a panel of 36 human IgG1 molecules with different variable region heavy/light chain pairings, but identical constant domains
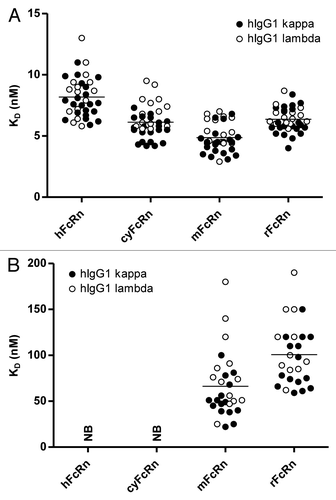
Wang et al.Citation23 examined the IgG-FcRn interaction of IgG with the same wild-type human Fc sequences, but different Fab domains. They found that the Fab domain, which has no direct contact with the FcRn, can affect the interaction with FcRn. However, the exact mechanism of how the Fab domain could affect this interaction is not yet clear. To further refine understanding of the influence of the Fab domain on FcRn binding, we used a unique IgG sample set, originally selected as scaffolds for the generation of our synthetic, fully human antibody phage display library Ylanthia®.Citation46 Ylanthia® is built on 36 fixed variable heavy (VH)/variable light (VL) chain pairs. These 36 VH/VL pairs comprise 12 different VH gene segments and 15 distinct VL gene segments (9 Vκ, 6 Vλ).Citation46 These 36 VH/VL pairs were expressed as full-length human IgG1 to examine the influence of the differences in the VH and VL gene segments (from N-terminus through framework 3, ) on the binding affinity to human, cynomolgus monkey, mouse and rat FcRn. Due to the size of the sample set, the above described BLI-based high throughput assay was used to analyze the IgG-FcRn interactions. As shown in , all 36 human IgGs showed very similar KD values at pH 6.0 on FcRn from the four different species. As expected there was no significant difference observed between kappa and lambda IgGs. Comparable to the previous analyses, FcRn binding at neutral pH was also examined. No binding was detectable for any of the 36 human IgG1 molecules to either human or cynomolgus monkey FcRn at pH 7.2, whereas binding was observed to mouse and rat FcRn ().
Figure 8. Comparison of determined equilibrium dissociation constants (KD) of interaction of 36 hIgG1 molecules with different VH/VL region combinationsCitation46 with FcRn from different species. Binding of 36 human variable domain IgG variants to human, cynomolgus monkey, mouse and rat FcRn at pH 6.0 (A) and pH 7.2 (B) was determined by BLI (see Materials and Methods for details). Mean KD values of samples are represented by the horizontal lines. KD values of 36 hIgG1 samples to the respective FcRn molecules were comparable at pH 6.0 (range between 3 and 13 nM). None of the 36 investigated hIgG1 samples bound to hFcRn or cyFcRn at neutral pH, whereas binding to mFcRn and rFcRn was observed (range between 25 and 190 nM). At neutral pH, evaluation was only possible for n = 28 (mFcRn) and n = 26 (rFcRn), respectively, due to weaker binding. As expected, no significant differences were observed between hIgG1 molecules consisting of kappa (filled circles) and lambda light chains (open circles), respectively. NB, no or negligible binding observed.
Discussion
In this study, we thoroughly investigated the interaction of different IgG isotypes and species orthologs to human, cynomolgus monkey, rat, and mouse FcRn. To the best of our knowledge, our study is the only one so far that analyzed the IgG-FcRn interaction of these four FcRn species in parallel.
Due to the high quality of the sample material (FcRn and IgG protein), which was generated with uniform protocols and production systems, as well as the consistent method setup and data evaluation, a valid comparison of the IgG-FcRn binding properties was possible. We consider the consistency of materials and experimental methods important because reported KD values for the IgG-FcRn interaction vary largely, most probably influenced by the different FcRn materials, assay formats and evaluation models that were used in the separate studies.
Beside these parameters, the absolute KD values can also vary due to different immobilization levels on the SPR sensor chip surface. As we observed, for generating robust results it is important to immobilize similar densities of active receptors on the sensor surfaces (unpublished observations). Magistrelli et al.Citation19 reported that amine coupling led to random orientation and reduced functionality of the FcRn molecules on the sensor chip surface. To avoid this drawback, we decided to capture biotinylated FcRn on a streptavidin coated chip surface as described by Magistrelli et al.Citation19 FcRn molecules can thus be conveniently immobilized at the desired surface density. Furthermore, this setup is probably a closer approximation to the in vivo situation of the receptor on the cell surface, as opposed to the reverse orientation (i.e., using the IgG as immobilized ligand and FcRn as the analyte). It is also more convenient for a characterization process in terms of higher throughput and lower material consumption.
In our experiments, a deviation of the resulting SPR sensorgrams from a monovalent interaction was observed (). This can be explained taking into account data from human and rat FcRn X-ray analysis of protein crystals, as well as analyses in solution, showing that two single FcRn molecules can interact with the Fc regions of one IgG heavy chain dimer.Citation12-Citation14 To describe the IgG-FcRn interaction, bivalent binding,Citation19,Citation42 two-state reaction,Citation33 heterogeneousCitation43,Citation50 or steady-state,Citation34,Citation39 as well as custom-developedCitation51 models have been published. The multiphasic interaction behavior makes it difficult to choose a correct model for determining and comparing KD values. In this study, the SPR and BLI data were fitted using the monovalent steady-state model, which generated apparent KD values due to the mentioned deviation from monovalent behavior. Compared with kinetic models, the steady-state evaluation model provided more robust results, and it was more suitable for a high throughput approach due to the much easier and faster generation of KD values.
In vitro FcRn binding characteristics of therapeutic IgG is considered indicative for their in vivo pharmacokinetic properties.Citation10,Citation23 Ideally, the interaction of IgG candidates with FcRn from different species should be analyzed already during early drug discovery in order to detect and exclude candidates with unusual or unfavorable binding characteristics. Though the current SPR-based method is suited and well-accepted to analyze FcRn interactions, it is highly beneficial to increase the throughput without compromising the quality of the generated results. The decreasing activity of the immobilized FcRn during long-term measurements limits the number of interactions that can be analyzed sequentially by SPR, and for a given platform the number of parallel interactions is usually fixed to the number of active flow cells. Therefore, we established a robust, high throughput BLI method allows fast sample analysis and fast evaluation of the results. We proved that this method is suitable for resolving even moderate differences in FcRn binding affinities by examining non-oxidized vs. oxidized IgG samples. When comparing results from the SPR- and the BLI-based methods, we observed moderate differences in the resulting absolute KD values.
It is important to point out that the absolute KD values can vary between both platforms due to the different experimental conditions. Probably the different FcRn immobilization levels on the SPR chip and BLI biosensor tip have the most important influence on the results. Due to the different detection principles, the FcRn immobilization levels cannot be easily transferred from SPR to BLI. Because of the lower sensitivity of the Octet system, higher immobilization levels are required to obtain appropriate binding signals. This might lead to partial mass-transport limitation during the measurement, which would be in line with higher affinities as determined by BLI in this study. However, immobilization levels can be controlled accurately enough to enable robust KD determination. In addition, both instruments allow a different level of temperature control: with Biacore, the temperature can be controlled independently of the ambient temperature, whereas in Octet, the plate temperature can only be kept constant above ambient temperature. The differences in experimental temperatures of 25 °C (SPR) or 30 °C (BLI) might also contribute to the observed affinity differences. As the relevance of moderate differences in KD in terms of in vivo half-life is unclear, we considered these differences in affinities as tolerable. Despite the differences in absolute KD values, the general binding patterns were well comparable. Consequently, we consider the BLI platform as suitable for IgG-FcRn interaction analysis in high throughput screening approaches.
In this study we examined the binding profile of the IgG-FcRn complex at both pH 6.0 and pH 7.2 independently. As shown by previous studies,Citation23,Citation52 the binding behavior at neutral pH is essential and seems to be potentially more important than binding at pH 6.0. Wang et al.Citation23 examined the IgG dissociation at neutral pH after binding at pH 6.0. They reported that IgGs with slower dissociation at neutral pH were associated with a shorter half-life in vivo.Citation23 The binding at neutral pH may reduce the possibility of the IgG to be released back into the blood stream. With the method reported here, an independent analysis at both pH values is possible, enabling a fast and robust evaluation of the IgG-FcRn binding profiles. If desired, candidates with suspicious binding profiles could also be investigated using the setup described by Wang et al.,Citation23 where FcRn binding at pH 6.0 and dissociation at pH 7.2 were analyzed in the same experiment. The described BLI-based method offers the flexibility to be easily adapted to follow this protocol, as sensor tips can be transferred from the pH 6.0 sample wells to those containing neutral pH buffer for dissociation of the IgG-FcRn complex. However, this setup has not yet been tested by us.
The pH-dependent binding of the FcRn-IgG complexes has been suggested to contribute to the IgG PK properties.Citation10,Citation23 Nevertheless, there are also published reports which suggest that multiple factors influence the PK of an IgG in vivo.Citation53,Citation54 Datta-Mannan et al.Citation53 presumed that both the characteristics of the IgG (e.g., biochemical and biophysical properties) and its therapeutic target (membrane-bound or soluble antigen, antigen load) can influence the disposition and elimination in vivo. In summary, a single in vitro parameter alone cannot readily predict in vivo PK, but FcRn binding is one important parameter that can influence the IgG PK in vivo. Consequently, it is essential to analyze this interaction in vitro already during early antibody discovery, e.g., by the approaches described in this report.
Using a larger panel of different antibodies than described in previous studies,Citation18,Citation23 our results clearly demonstrate that IgG isotypes have a negligible influence on FcRn binding. Significant differences in the IgG-FcRn binding pattern were only observed for binding of IgG molecules to the different tested rodent and primate FcRn species.
Wang et al.Citation23 suggested that the Fab domain may affect the FcRn interaction. To further examine the effect, we analyzed 36 different VH/VL variable region combinations. It was observed that the analyzed VH/VL region pairings (representing all combinations implemented in the Ylanthia® library) were “well-behaved”, i.e., showed the typical binding properties of human IgG molecules. However, it has been reported, that anomalies in FcRn binding can occur with human antibodies, e.g., increased binding at neutral pH.Citation35,Citation39 Our results clearly show that variations in the framework parts or the CDR1 or CDR2 of the Fab domain (i.e., other VH/VL region combinations) did not significantly change FcRn binding. It should be noted, however, that all of the tested variable regions were of human germline composition.Citation46 Thus, it cannot be ruled out that the use of “unnatural” CDR and framework residues or sequences might finally lead to the differences observed by Wang et al. In addition, different CDR3 sequences might affect the FcRn binding. As the CDR3s of heavy and light chain are the CDRs that lie closest to the FcRn binding site,Citation11 this assumption would also make a relatively important contribution to FcRn binding plausible.
Especially in the context of a likely influence of the CDRs on FcRn binding, we consider it indispensable to assess the FcRn binding for each individual therapeutic antibody candidate. Depending on the project stage, this demands a robust method capable of handling large number of candidate molecules. We are convinced that the BLI-based method described here is a valuable tool for analyzing the pH dependent IgG-FcRn binding properties. Thus, it will facilitate the selection of potent antibody candidates in early discovery phases and thereby increase the probability of success in later development stages.
Materials and Methods
Construction of FcRn molecules
The cDNAs encoding the human (GenBank accession number P55899), cynomolgus monkey (Q8SPV9), mouse (Q61559) and rat (P13599) FcRn α-chain, also called p51, as well as the cDNAs encoding the human (P61769), cynomolgus monkey (Q8SPW0), mouse (P01887) and rat (P07151) β2m, also named p14, were gene-optimized and synthesized using the Slonomics® technology.Citation55
The extracellular domains of the FcRn α-chain were amplified by PCR. Amino acids 1–297 were used for human, cynomolgus and mouse FcRn α-chain, while amino acids 1–298 were amplified for the rat FcRn α-chain. After restriction digest, the PCR fragments were inserted into a pMAX vector in frame with a C-terminal avi-6xHis tag creating the pMAX_p51_avi-H expression vectors. The pMAX vector series is based on a modified pcDNA3 (Life Technologies) vector enabling the subcloning of cDNAs in frame with various tags. Accordingly, the whole sequences of the β2m were subcloned into pMAX vectors resulting in the pMAX_p14 expression vectors.
Subsequently, the expression cassettes comprising the CMV promotor sequence, the ORF of the β2m and the polyadenylation site, were subcloned into the BglII restriction site of the pMAX_p51_avi-H vectors generating the final one-vector expression constructs pMAX_p51_avi-H/p14.
Construction of immunoglobulin molecules
The anti-lysozyme antibody MOR03207 was selected using Human Combinatorial Antibody Library (HuCAL) GOLD®.Citation56 To obtain full-length IgG molecules of different isotypes and species (human IgG1, 2, 4, 4_S241P; cynomolgus monkey IgG1, 2, 4; mouse IgG2a; rat IgG2a), the variable domains of the light and the heavy chain were sub-cloned into pMorph2 expression vectors encoding the respective constant region of the IgG species and isotype (light chain: pMorph2_h_Iglambda2, pMorph2_h/cy_Iglambda, pMorph2_h/m_Iglambda, pMorph2_h/r_Iglambda; heavy chain: pMorph_h_IgG1f, pMorph2_h_IgG2, pMorph2_h_IgG4, pMorph2_h_IgG4_S241P, pMorph2_h/cy_IgG1, pMorph2_h/cy_IgG2, pMorph2_h/cy_IgG4, pMorph2_h/m_IgG2a, pMorph2_h/r_IgG2a).
Mutation of serine 241 (numbering according to Kabat) to proline in the hinge region (C-P-S-C) of the human IgG4 molecule (IgG4_S241P) was performed to reduce formation of ‘half-molecules’.Citation57
The 36 V region pairs that were used for the generation of the Ylanthia® IgG1 scaffolds were composed of 12 different human Ig variable heavy chain regions (VH1–18, VH1–46, VH1–69, VH3–07, VH3–11, VH3–15, VH3–21, VH3–23, VH3–53, VH3–74, VH5–51, VH6–01) and 15 distinct human Ig variable light chain regions (Vκ1–05, Vκ1–06, Vκ1–09, Vκ1–12, Vκ1–27, Vκ1–39, Vκ3–11, Vκ3–15, Vκ3–20, Vλ1–40, Vλ1–47, Vλ1–51, Vλ2–11, Vλ2–23, Vλ3–1). All VH regions carried identical CDR-H3s of the hu4D5–8 antibodyCitation58,Citation59 (WGGDGFYAMDY) and the JH4 joining segment. The VL regions carried either a kappa-like CDR-L3 (QQHYTTPPT, of hu4D5–8 antibody) and Jκ1 joining segment for kappa light chains, or a lambda-like CDR-L3 (QSYDSSLSGVV) and the Jλ2/3 joining segment for lambda light chains.Citation60
The variable domains of the heavy chain and light chain were cloned into the pYMex20_h_IgG1f expression vectors as described by Tiller et al.Citation46
Transient transfection and protein purification
Eukaryotic HKB11 cellsCitation61 were transiently transfected using lipofectamine 2000 (Life Technologies) or polyethylenimine (Polyplus). The cell culture supernatants were harvested three or six days post-transfection.
After sterile filtration, the recombinant human, cynomolgus monkey, mouse and rat FcRn molecules were purified via immobilized metal ion affinity chromatography (IMAC, Protino® Ni-TED, Macherey-Nagel). Buffer exchange to DPBS (Life Technologies, 14190) was done using PD10 gel filtration columns (GE Healthcare) and the samples were finally sterile filtered (0.2 µm pore size). Purified protein concentrations were determined by UV spectrophotometry (Thermo Scientific). Expression yields were calculated as amount of IMAC purified FcRn per liter cell culture supernatant.
Accordingly, human, cynomolgus monkey, and mouse IgGs were purified via Protein A affinity chromatography (MabSelect SURE, GE Healthcare), while rat IgGs were subjected to Gamma bind affinity chromatography (GE Healthcare). Buffer exchange to DPBS and sterile filtration were performed as described above for the FcRn preparation.
Characterization of purified materials
FcRn and IgG materials were analyzed under denaturing, reducing and denaturing, non-reducing conditions by SDS-PAGE (NuPAGE, Life Technologies) and Coomassie Blue staining (Thermo Scientific) or by capillary electrophoresis (LabChip® GX II, Perkin Elmer).
The monomer content of the heterodimeric FcRn and the heterotetrameric IgG molecules was determined by high performance size exclusion chromatography (HP-SEC) applying a TSKgel G3000SWxl column (Tosoh Bioscience GmbH) on an UltiMate® 3000 Titanium HPLC system (Dionex/Thermo Scientific). The system was used in line with miniDAWNTM Treos® and Optilab® rEX detectors (Wyatt Technology) for absolute molar mass determination. For each sample, 15 µg (FcRn) or 7.5 µg (IgG) of protein were injected onto the SEC column. Separation was performed at a flow rate of 0.5 mL/min using 150 mM potassium phosphate, pH 6.5, as a mobile phase. UV absorbance at 280 nm (FcRn) or 210 nm (IgG) was recorded.
Biotinylation
Biotinylation was performed using biotin protein ligase (Avidity) according to the manufacturer’s instructions. After biotinylation, the free biotin was removed using PD10 gel filtration columns.
FcRn binding characterization using SPR
SPR experiments were conducted at 25 °C using a Biacore T200 instrument (GE Healthcare). Purified and biotinylated FcRn was immobilized on a Biacore Streptavidin biosensor chip (GE Healthcare, BR-1005–31) as described by the manufacturer. An immobilization level of ~200 response units (RU) was reached.
For kinetic experiments, IgG samples were diluted to concentrations ranging from 3000 nM to 4 nM with assay buffer (DPBS (Life Technologies, 14190), adjusted to pH 6.0 with HCl, 0.05% (v/v) Tween 20 (Merck, 8.22184.0500)). The solutions were injected sequentially over flow cells immobilized with human, cynomolgus monkey, mouse and rat FcRn or assay buffer (see above) as a blank. The association phase was set to 240 s and dissociation was 240 s. At the end of the dissociation phase, HBS-EP+, adjusted to pH 8.0 (GE Healthcare, BR-1006–69) was injected two times for 30 s to regenerate the sensor surface. The experiments at neutral pH were performed as described above except that an assay buffer was applied without pH adjustment (pH 7.2) for sample dilution and as running buffer.
The sensorgram data was processed by subtracting the signals obtained from the uncoated surface and from a blank injection of assay buffer (double referencing). Binding analysis was performed using the Biacore T200 Evaluation Software Version 1.0 (GE Healthcare). The sensorgrams and steady-state fits were plotted using Microsoft Excel 2010. The following fit model was used:
with Y, binding signal (RU); Rmax, maximum binding level observed (RU); x, the IgG concentration (M); KD, the equilibrium dissociation constant (M).
FcRn binding characterization using BLI
BLI experiments were performed on an Octet QK384 (PALL/ForteBio) instrument at 30 °C. Streptavidin coated biosensors (PALL/ForteBio, 18–5021) were loaded with purified and biotinylated human, cynomolgus monkey, mouse or rat FcRn in assay buffer (DPBS (Life Technologies, 14190), adjusted to pH 6.0 with HCl, 0.05% (v/v) Tween 20 (Merck, 8.22184.0500)). Immobilization levels between 0.6 nm and 1.0 nm were reached.
For association phase monitoring, IgG samples were diluted with assay buffer pH 6.0 to concentrations ranging from 1000 nM to 0.5 nM and transferred to solid-black 384 well plates (Greiner Bio-One, 781900). IgG samples were allowed to bind to FcRn loaded biosensors for 300 s. The dissociation phase was recorded in wells of a solid black 96-well plate (Greiner Bio-One, 655900) containing assay buffer pH 6.0 for 150 s. The biosensors were regenerated two times with HBS-EP+ pH 8.0 (GE Healthcare, BR-1006–69) for 30 s. Between regeneration steps, biosensors were washed with pH 6.0 assay buffer for 20 s.
The experiments at neutral pH were performed as described above except that an assay buffer with pH 7.2 was used for sample dilution, dissociation and intermittent washing steps.
All data were referenced with FcRn loaded Streptavidin-coated biosensors incubated in assay buffer instead of analyte. The sensorgrams were plotted and evaluated using data analysis software version 7.0 (PALL/ForteBio). The steady-state fits were plotted using Microsoft Excel 2010.
| Abbreviations: | ||
| BLI | = | biolayer interferometry |
| CDR | = | complementarity-determining region |
| cyFcRn | = | cynomolgus monkey FcRn |
| DLS | = | dynamic light scattering |
| Fab | = | fragment antigen binding |
| Fc | = | fragment crystallizable |
| FcRn | = | neonatal Fc receptor |
| β2m | = | β2 microglobulin |
| hFcRn | = | human FcRn |
| HuCAL® | = | human combinatorial antibody library |
| IgG | = | immunoglobulin G |
| KD | = | equilibrium dissociation constant |
| mFcRn | = | mouse FcRn |
| PBS | = | phosphate buffered saline |
| PK | = | pharmacokinetics |
| rFcRn | = | rat FcRn |
| RT | = | room temperature |
| RU | = | resonance unit(s) |
| SPR | = | surface plasmon resonance |
| VH | = | variable Ig heavy chain region |
| VL | = | variable Ig light chain region |
Additional material
Download Zip (5.8 MB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank MorphoSys’ Physico-Chemical Analytics for characterization of Fc receptors, Martin Heßling for contribution of ideas, Stefanie Urlinger and Stefan Härtle for helpful comments and reviewing of the manuscript.
Trademarks Disclosure
HuCAL®, HuCAL GOLD® and Ylanthia® are registered trademarks of MorphoSys AG. Slonomics® is a registered trademark of Sloning BioTechnology GmbH, a subsidiary of MorphoSys AG.
References
- Reichert JM. Antibodies to watch in 2014. MAbs 2013; 6:5 - 14; PMID: 24284914
- Reichert JM. Which are the antibodies to watch in 2013?. MAbs 2013; 5:1 - 4; http://dx.doi.org/10.4161/mabs.22976; PMID: 23254906
- Bumbaca D, Boswell CA, Fielder PJ, Khawli LA. Physiochemical and biochemical factors influencing the pharmacokinetics of antibody therapeutics. AAPS J 2012; 14:554 - 8; http://dx.doi.org/10.1208/s12248-012-9369-y; PMID: 22610647
- Roopenian DC, Akilesh S. FcRn: the neonatal Fc receptor comes of age. Nat Rev Immunol 2007; 7:715 - 25; http://dx.doi.org/10.1038/nri2155; PMID: 17703228
- Ghetie V, Ward ES. Multiple roles for the major histocompatibility complex class I- related receptor FcRn. Annu Rev Immunol 2000; 18:739 - 66; http://dx.doi.org/10.1146/annurev.immunol.18.1.739; PMID: 10837074
- Kacskovics I, Cervenak J, Erdei A, Goldsby RA, Butler JE. Recent advances using FcRn overexpression in transgenic animals to overcome impediments of standard antibody technologies to improve the generation of specific antibodies. MAbs 2011; 3:431 - 9; http://dx.doi.org/10.4161/mabs.3.5.17023; PMID: 22048692
- Anderson CL, Chaudhury C, Kim J, Bronson CL, Wani MA, Mohanty S. Perspective-- FcRn transports albumin: relevance to immunology and medicine. Trends Immunol 2006; 27:343 - 8; http://dx.doi.org/10.1016/j.it.2006.05.004; PMID: 16731041
- Kim J, Bronson CL, Hayton WL, Radmacher MD, Roopenian DC, Robinson JM, Anderson CL. Albumin turnover: FcRn-mediated recycling saves as much albumin from degradation as the liver produces. Am J Physiol Gastrointest Liver Physiol 2006; 290:G352 - 60; http://dx.doi.org/10.1152/ajpgi.00286.2005; PMID: 16210471
- Zalevsky J, Chamberlain AK, Horton HM, Karki S, Leung IW, Sproule TJ, Lazar GA, Roopenian DC, Desjarlais JR. Enhanced antibody half-life improves in vivo activity. Nat Biotechnol 2010; 28:157 - 9; http://dx.doi.org/10.1038/nbt.1601; PMID: 20081867
- Kuo TT, Aveson VG. Neonatal Fc receptor and IgG-based therapeutics. MAbs 2011; 3:422 - 30; http://dx.doi.org/10.4161/mabs.3.5.16983; PMID: 22048693
- Rodewald R. pH-dependent binding of immunoglobulins to intestinal cells of the neonatal rat. J Cell Biol 1976; 71:666 - 9; http://dx.doi.org/10.1083/jcb.71.2.666; PMID: 11223
- Martin WL, West AP Jr., Gan L, Bjorkman PJ. Crystal structure at 2.8 A of an FcRn/heterodimeric Fc complex: mechanism of pH-dependent binding. Mol Cell 2001; 7:867 - 77; http://dx.doi.org/10.1016/S1097-2765(01)00230-1; PMID: 11336709
- Burmeister WP, Huber AH, Bjorkman PJ. Crystal structure of the complex of rat neonatal Fc receptor with Fc. Nature 1994; 372:379 - 83; http://dx.doi.org/10.1038/372379a0; PMID: 7969498
- West AP Jr., Bjorkman PJ. Crystal structure and immunoglobulin G binding properties of the human major histocompatibility complex-related Fc receptor(,). Biochemistry 2000; 39:9698 - 708; http://dx.doi.org/10.1021/bi000749m; PMID: 10933786
- Shields RL, Namenuk AK, Hong K, Meng YG, Rae J, Briggs J, Xie D, Lai J, Stadlen A, Li B, et al. High resolution mapping of the binding site on human IgG1 for Fc gamma RI, Fc gamma RII, Fc gamma RIII, and FcRn and design of IgG1 variants with improved binding to the Fc gamma R. J Biol Chem 2001; 276:6591 - 604; http://dx.doi.org/10.1074/jbc.M009483200; PMID: 11096108
- Kim JK, Firan M, Radu CG, Kim CH, Ghetie V, Ward ES. Mapping the site on human IgG for binding of the MHC class I-related receptor, FcRn. Eur J Immunol 1999; 29:2819 - 25; http://dx.doi.org/10.1002/(SICI)1521-4141(199909)29:09<2819::AID-IMMU2819>3.0.CO;2-6; PMID: 10508256
- Vaccaro C, Bawdon R, Wanjie S, Ober RJ, Ward ES. Divergent activities of an engineered antibody in murine and human systems have implications for therapeutic antibodies. Proc Natl Acad Sci U S A 2006; 103:18709 - 14; http://dx.doi.org/10.1073/pnas.0606304103; PMID: 17116867
- Ober RJ, Radu CG, Ghetie V, Ward ES. Differences in promiscuity for antibody-FcRn interactions across species: implications for therapeutic antibodies. Int Immunol 2001; 13:1551 - 9; http://dx.doi.org/10.1093/intimm/13.12.1551; PMID: 11717196
- Magistrelli G, Malinge P, Anceriz N, Desmurs M, Venet S, Calloud S, Daubeuf B, Kosco-Vilbois M, Fischer N. Robust recombinant FcRn production in mammalian cells enabling oriented immobilization for IgG binding studies. J Immunol Methods 2012; 375:20 - 9; http://dx.doi.org/10.1016/j.jim.2011.09.002; PMID: 21939661
- Zhou J, Mateos F, Ober RJ, Ward ES. Conferring the binding properties of the mouse MHC class I-related receptor, FcRn, onto the human ortholog by sequential rounds of site-directed mutagenesis. J Mol Biol 2005; 345:1071 - 81; http://dx.doi.org/10.1016/j.jmb.2004.11.014; PMID: 15644205
- Andersen JT, Daba MB, Berntzen G, Michaelsen TE, Sandlie I. Cross-species binding analyses of mouse and human neonatal Fc receptor (FcRn) show dramatic differences in immunoglobulin G (IgG) and albumin binding. J Biol Chem 2010; 285:4826 - 36; http://dx.doi.org/10.1074/jbc.M109.081828
- Tam SH, McCarthy SG, Brosnan K, Goldberg KM, Scallon BJ. Correlations between pharmacokinetics of IgG antibodies in primates vs. FcRn-transgenic mice reveal a rodent model with predictive capabilities. MAbs 2013; 5:1 - 9; http://dx.doi.org/10.4161/mabs.23836; PMID: 23549129
- Wang W, Lu P, Fang Y, Hamuro L, Pittman T, Carr B, Hochman J, Prueksaritanont T. Monoclonal antibodies with identical Fc sequences can bind to FcRn differentially with pharmacokinetic consequences. Drug Metab Dispos 2011; 39:1469 - 77; http://dx.doi.org/10.1124/dmd.111.039453; PMID: 21610128
- Andersen JT, Foss S, Kenanova VE, Olafsen T, Leikfoss IS, Roopenian DC, Wu AM, Sandlie I. Anti-carcinoembryonic antigen single-chain variable fragment antibody variants bind mouse and human neonatal Fc receptor with different affinities that reveal distinct cross-species differences in serum half-life. J Biol Chem 2012; 287:22927 - 37; http://dx.doi.org/10.1074/jbc.M112.355131; PMID: 22570488
- Yeung YA, Leabman MK, Marvin JS, Qiu J, Adams CW, Lien S, Starovasnik MA, Lowman HB. Engineering human IgG1 affinity to human neonatal Fc receptor: impact of affinity improvement on pharmacokinetics in primates. J Immunol 2009; 182:7663 - 71; http://dx.doi.org/10.4049/jimmunol.0804182; PMID: 19494290
- Martin WL, Bjorkman PJ. Characterization of the 2:1 complex between the class I MHC-related Fc receptor and its Fc ligand in solution. Biochemistry 1999; 38:12639 - 47; http://dx.doi.org/10.1021/bi9913505; PMID: 10504233
- Igawa T, Tsunoda H, Tachibana T, Maeda A, Mimoto F, Moriyama C, Nanami M, Sekimori Y, Nabuchi Y, Aso Y, et al. Reduced elimination of IgG antibodies by engineering the variable region. Protein Eng Des Sel 2010; 23:385 - 92; http://dx.doi.org/10.1093/protein/gzq009; PMID: 20159773
- Andersen JT, Justesen S, Berntzen G, Michaelsen TE, Lauvrak V, Fleckenstein B, Buus S, Sandlie I. A strategy for bacterial production of a soluble functional human neonatal Fc receptor. J Immunol Methods 2008; 331:39 - 49; http://dx.doi.org/10.1016/j.jim.2007.11.003; PMID: 18155020
- Lu Y, Vernes JM, Chiang N, Ou Q, Ding J, Adams C, Hong K, Truong BT, Ng D, Shen A, et al. Identification of IgG(1) variants with increased affinity to FcγRIIIa and unaltered affinity to FcγRI and FcRn: comparison of soluble receptor-based and cell-based binding assays. J Immunol Methods 2011; 365:132 - 41; http://dx.doi.org/10.1016/j.jim.2010.12.014; PMID: 21185301
- Kelley RF, Meng YG. Methods to engineer and identify IgG1 variants with improved FcRn binding or effector function. Methods Mol Biol 2012; 901:277 - 93; http://dx.doi.org/10.1007/978-1-61779-931-0_18; PMID: 22723108
- Hinton PR, Xiong JM, Johlfs MG, Tang MT, Keller S, Tsurushita N. An engineered human IgG1 antibody with longer serum half-life. J Immunol 2006; 176:346 - 56; PMID: 16365427
- Mathur A, Arora T, Liu L, Crouse-Zeineddini J, Mukku V. Qualification of a homogeneous cell-based neonatal Fc receptor (FcRn) binding assay and its application to studies on Fc functionality of IgG-based therapeutics. J Immunol Methods 2013; 390:81 - 91; http://dx.doi.org/10.1016/j.jim.2013.01.011; PMID: 23384837
- Bertolotti-Ciarlet A, Wang W, Lownes R, Pristatsky P, Fang Y, McKelvey T, Li Y, Li Y, Drummond J, Prueksaritanont T, et al. Impact of methionine oxidation on the binding of human IgG1 to Fc Rn and Fc gamma receptors. Mol Immunol 2009; 46:1878 - 82; http://dx.doi.org/10.1016/j.molimm.2009.02.002; PMID: 19269032
- Stapleton NM, Andersen JT, Stemerding AM, Bjarnarson SP, Verheul RC, Gerritsen J, Zhao Y, Kleijer M, Sandlie I, de Haas M, et al. Competition for FcRn-mediated transport gives rise to short half-life of human IgG3 and offers therapeutic potential. Nat Commun 2011; 2:599; http://dx.doi.org/10.1038/ncomms1608; PMID: 22186895
- Dall’Acqua WF, Woods RM, Ward ES, Palaszynski SR, Patel NK, Brewah YA, Wu H, Kiener PA, Langermann S. Increasing the affinity of a human IgG1 for the neonatal Fc receptor: biological consequences. J Immunol 2002; 169:5171 - 80; PMID: 12391234
- Dall’Acqua WF, Kiener PA, Wu H. Properties of human IgG1s engineered for enhanced binding to the neonatal Fc receptor (FcRn). J Biol Chem 2006; 281:23514 - 24; http://dx.doi.org/10.1074/jbc.M604292200; PMID: 16793771
- Datta-Mannan A, Witcher DR, Tang Y, Watkins J, Jiang W, Wroblewski VJ. Humanized IgG1 variants with differential binding properties to the neonatal Fc receptor: relationship to pharmacokinetics in mice and primates. Drug Metab Dispos 2007; 35:86 - 94; http://dx.doi.org/10.1124/dmd.106.011734; PMID: 17050651
- Presta LG. Molecular engineering and design of therapeutic antibodies. Curr Opin Immunol 2008; 20:460 - 70; http://dx.doi.org/10.1016/j.coi.2008.06.012; PMID: 18656541
- Deng R, Loyet KM, Lien S, Iyer S, DeForge LE, Theil FP, Lowman HB, Fielder PJ, Prabhu S. Pharmacokinetics of humanized monoclonal anti-tumor necrosis factor-alpha antibody and its neonatal Fc receptor variants in mice and cynomolgus monkeys. Drug Metab Dispos 2010; 38:600 - 5; http://dx.doi.org/10.1124/dmd.109.031310; PMID: 20071453
- Waldmann TA, Strober W. Metabolism of immunoglobulins. Prog Allergy 1969; 13:1 - 110; PMID: 4186070
- Gandhi M, Alwawi E, Gordon KB. Anti-p40 antibodies ustekinumab and briakinumab: blockade of interleukin-12 and interleukin-23 in the treatment of psoriasis. Semin Cutan Med Surg 2010; 29:48 - 52; http://dx.doi.org/10.1016/j.sder.2010.02.001; PMID: 20430307
- Suzuki T, Ishii-Watabe A, Tada M, Kobayashi T, Kanayasu-Toyoda T, Kawanishi T, Yamaguchi T. Importance of neonatal FcR in regulating the serum half-life of therapeutic proteins containing the Fc domain of human IgG1: a comparative study of the affinity of monoclonal antibodies and Fc-fusion proteins to human neonatal FcR. J Immunol 2010; 184:1968 - 76; http://dx.doi.org/10.4049/jimmunol.0903296; PMID: 20083659
- Datta-Mannan A, Witcher DR, Tang Y, Watkins J, Wroblewski VJ. Monoclonal antibody clearance. Impact of modulating the interaction of IgG with the neonatal Fc receptor. J Biol Chem 2007; 282:1709 - 17; http://dx.doi.org/10.1074/jbc.M607161200; PMID: 17135257
- Igawa T, Ishii S, Tachibana T, Maeda A, Higuchi Y, Shimaoka S, Moriyama C, Watanabe T, Takubo R, Doi Y, et al. Antibody recycling by engineered pH-dependent antigen binding improves the duration of antigen neutralization. Nat Biotechnol 2010; 28:1203 - 7; http://dx.doi.org/10.1038/nbt.1691; PMID: 20953198
- Igawa T, Tsunoda H, Kuramochi T, Sampei Z, Ishii S, Hattori K. Engineering the variable region of therapeutic IgG antibodies. MAbs 2011; 3:243 - 52; http://dx.doi.org/10.4161/mabs.3.3.15234; PMID: 21406966
- Tiller T, Schuster I, Deppe D, Siegers K, Strohner R, Herrmann T, Berenguer M, Poujol D, Stehle J, Stark Y, et al. A fully synthetic human Fab antibody library based on fixed VH/VL framework pairings with favorable biophysical properties. MAbs 2013; 5:1 - 26; http://dx.doi.org/10.4161/mabs.24218; PMID: 23571156
- Abdiche Y, Malashock D, Pinkerton A, Pons J. Determining kinetics and affinities of protein interactions using a parallel real-time label-free biosensor, the Octet. Anal Biochem 2008; 377:209 - 17; http://dx.doi.org/10.1016/j.ab.2008.03.035; PMID: 18405656
- Wang W, Vlasak J, Li Y, Pristatsky P, Fang Y, Pittman T, Roman J, Wang Y, Prueksaritanont T, Ionescu R. Impact of methionine oxidation in human IgG1 Fc on serum half-life of monoclonal antibodies. Mol Immunol 2011; 48:860 - 6; http://dx.doi.org/10.1016/j.molimm.2010.12.009; PMID: 21256596
- Liu D, Ren D, Huang H, Dankberg J, Rosenfeld R, Cocco MJ, Li L, Brems DN, Remmele RL Jr.. Structure and stability changes of human IgG1 Fc as a consequence of methionine oxidation. Biochemistry 2008; 47:5088 - 100; http://dx.doi.org/10.1021/bi702238b; PMID: 18407665
- Andersen JT, Dee Qian J, Sandlie I. The conserved histidine 166 residue of the human neonatal Fc receptor heavy chain is critical for the pH-dependent binding to albumin. Eur J Immunol 2006; 36:3044 - 51; http://dx.doi.org/10.1002/eji.200636556; PMID: 17048273
- Gurbaxani BM, Morrison SL. Development of new models for the analysis of Fc-FcRn interactions. Mol Immunol 2006; 43:1379 - 89; http://dx.doi.org/10.1016/j.molimm.2005.08.002; PMID: 16183124
- Vaccaro C, Zhou J, Ober RJ, Ward ES. Engineering the Fc region of immunoglobulin G to modulate in vivo antibody levels. Nat Biotechnol 2005; 23:1283 - 8; http://dx.doi.org/10.1038/nbt1143; PMID: 16186811
- Datta-Mannan A, Chow CK, Dickinson C, Driver D, Lu J, Witcher DR, Wroblewski VJ. FcRn affinity-pharmacokinetic relationship of five human IgG4 antibodies engineered for improved in vitro FcRn binding properties in cynomolgus monkeys. Drug Metab Dispos 2012; 40:1545 - 55; http://dx.doi.org/10.1124/dmd.112.045864; PMID: 22584253
- Hötzel I, Theil FP, Bernstein LJ, Prabhu S, Deng R, Quintana L, Lutman J, Sibia R, Chan P, Bumbaca D, et al. A strategy for risk mitigation of antibodies with fast clearance. MAbs 2012; 4:753 - 60; http://dx.doi.org/10.4161/mabs.22189; PMID: 23778268
- Van den Brulle J, Fischer M, Langmann T, Horn G, Waldmann T, Arnold S, Fuhrmann M, Schatz O, O’Connell T, O’Connell D, et al. A novel solid phase technology for high-throughput gene synthesis. Biotechniques 2008; 45:340 - 3; http://dx.doi.org/10.2144/000112953; PMID: 18778261
- Rothe C, Urlinger S, Löhning C, Prassler J, Stark Y, Jäger U, Hubner B, Bardroff M, Pradel I, Boss M, et al. The human combinatorial antibody library HuCAL GOLD combines diversification of all six CDRs according to the natural immune system with a novel display method for efficient selection of high-affinity antibodies. J Mol Biol 2008; 376:1182 - 200; http://dx.doi.org/10.1016/j.jmb.2007.12.018; PMID: 18191144
- Schuurman J, Perdok GJ, Gorter AD, Aalberse RC. The inter-heavy chain disulfide bonds of IgG4 are in equilibrium with intra-chain disulfide bonds. Mol Immunol 2001; 38:1 - 8; http://dx.doi.org/10.1016/S0161-5890(01)00050-5; PMID: 11483205
- Knappik A, Ge L, Honegger A, Pack P, Fischer M, Wellnhofer G, Hoess A, Wölle J, Plückthun A, Virnekäs B. Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and CDRs randomized with trinucleotides. J Mol Biol 2000; 296:57 - 86; http://dx.doi.org/10.1006/jmbi.1999.3444; PMID: 10656818
- Carter P, Presta L, Gorman CM, Ridgway JB, Henner D, Wong WL, Rowland AM, Kotts C, Carver ME, Shepard HM. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc Natl Acad Sci U S A 1992; 89:4285 - 9; http://dx.doi.org/10.1073/pnas.89.10.4285; PMID: 1350088
- Ewert S, Huber T, Honegger A, Plückthun A. Biophysical properties of human antibody variable domains. J Mol Biol 2003; 325:531 - 53; http://dx.doi.org/10.1016/S0022-2836(02)01237-8; PMID: 12498801
- Cho MS, Yee H, Chan S. Establishment of a human somatic hybrid cell line for recombinant protein production. J Biomed Sci 2002; 9:631 - 8; http://dx.doi.org/10.1007/BF02254991; PMID: 12432229
