Abstract
HER2, a ligand-free tyrosine kinase receptor of the HER family, is frequently overexpressed in breast cancer. The anti-HER2 antibody trastuzumab has shown significant clinical benefits in metastatic breast cancer; however, resistance to trastuzumab is common. The development of monoclonal antibodies that have complementary mechanisms of action results in a more comprehensive blockade of ErbB2 signaling, especially HER2/HER3 signaling. Use of such antibodies may have clinical benefits if these antibodies can become widely accepted. Here, we describe a novel anti-HER2 antibody, hHERmAb-F0178C1, which was isolated from a screen of a phage display library. A step-by-step optimization method was employed to maximize the inhibitory effect of this anti-HER2 antibody. Crystallographic analysis was used to determine the three-dimensional structure to 3.5 Å resolution, confirming that the epitope of this antibody is in domain III of HER2. Moreover, this novel anti-HER2 antibody exhibits superior efficacy in blocking HER2/HER3 heterodimerization and signaling, and its use in combination with pertuzumab has a synergistic effect. Characterization of this antibody revealed the important role of a ligand binding site within domain III of HER2. The results of this study clearly indicate the unique potential of hHERmAb-F0178C1, and its complementary inhibition effect on HER2/HER3 signaling warrants its consideration as a promising clinical treatment.
Introduction
Amplification of the human epidermal growth factor receptor 2 (HER2) gene, and therefore the ErbB2 receptor, occurs in ~20–30% of breast tumors and is associated with a more malignant phenotype, resistance to chemotherapy, and poor prognosis.Citation1-Citation4 However, unlike other ErbB receptors, HER2 has no known ligand.Citation5 To date, there are at least 11 different EGF family ligands that bind to HER receptors, including epidermal growth factor (EGF), transforming growth factor α (TGF-α), and neuregulins (NRGs).Citation6,Citation7 The binding of these ligands to their cognate receptors results in the dimerization and activation of the receptor, which transmits downstream growth signals. Although no ligand has been reported that directly binds to HER2, it is considered to be an important coreceptor partner of other HERs. The importance of HER2 has led to the development of targeted therapies that are tumor-selective and effective for extending life expectancy in patients with early and advanced breast cancer, such as trastuzumab (Herceptin®), a humanized monoclonal IgG1 that binds to the ectodomain of HER2, induces clinical responses in HER2-overexpressing breast cancers, and prolongs patient survival when combined with chemotherapy.Citation8 This antibody is now in clinical use for metastatic breast cancer overexpressing ErbB2, but up to 70% of patients are resistant to the treatment.Citation9 The clinical efficacy of trastuzumab appears to be limited to breast cancers that overexpress HER2, as measured by intense membrane staining in the majority of tumor cells with HER2 antibodies (3+ by immunohistochemistry) or excess copies of the HER2 gene determined by fluorescence in situ hybridization. Therefore, HER2 overexpression as detected by immunohistochemistry or fluorescence in situ hybridization is a biomarker that is predictive of a good response to treatment with the antibody. However, many patients with HER2 gene-amplified metastatic breast cancers do not respond to or eventually escape trastuzumab’s effects, suggesting both de novo and acquired mechanisms of therapeutic resistance. Nevertheless, researchers are especially interested in pertuzumab (Perjeta®) for the treatment of HER2-positive breast cancer since its approval by the US Food and Drug Administration in 2012. Pertuzumab is an antibody that binds to the dimerization arm of ErbB2 and prevents its function as a coreceptor with the other ErbBs, most notably ErbB3.Citation10,Citation11 The Phase 3 trial that supported the approval of pertuzumabCitation12 was conducted in 808 patients with metastatic breast cancer receiving first-line treatment. The individuals in the treatment group who received the combination of pertuzumab, trastuzumab, and docetaxel showed a significant reduction in the risk of progression or death and an absolute increase in progression-free survival of 6.1 mo. Notably, recent reports suggest that trastuzumab does not block ErbB2 activation, but rather disrupts noncanonical ErbB2–ErbB3 interactions that result in constitutive ErbB3 activation.Citation13 Importantly, the aforementioned clinical results with pertuzumab and trastuzumab validate the laboratory findings. Taken together, these findings suggest that targeting ErbB2 with monoclonal antibodies (mAbs) that have complementary mechanisms of action results in a more comprehensive blockade of ErbB2 signaling. Thus, the development of novel antibodies that may have complementary blocking effects on HER2 is urgently needed.
With the aim of developing a novel therapeutic agent for HER2 signaling inhibition, we developed a novel anti-HER2 antibody, hHERmAb-F0178, which binds to domain III of HER2 and was enhanced using a step-by-step antibody optimization method. The crystal structure of human HER2 complexed with the final antibody was solved at 3.5 Å resolution to confirm the epitope of this antibody. This novel anti-HER2 antibody exhibits superior efficacy for blocking HER2/HER3 heterodimerization and signaling, and its use in combination with pertuzumab has a synergistic effect. The characterization of this novel antibody reveals the important role of ligand binding domain III of HER2.
Results
ErbB2-specific mAbs cloned from human B cell phage display libraries
The previously described large non-immunized repertoires of human scFv fragments displayed on phageCitation14,Citation15 were used for the selection of single chain antibodies that bound to the human HER2 extracellular domain (ECD) protein. More than 800 distinct HER2-binding scFv fragments were identified and were ranked based on their affinity for binding to recombinant HER2 ECD protein. A panel of 25 candidate scFv fragments that demonstrated the greatest binding affinity and did not compete with trastuzumab and pertuzumab for binding to HER2 were selected for further analysis. These scFv fragments were converted to full IgG molecules (IgG1,κ) and assessed for their ability to inhibit the proliferation of BT474 and MCF-7 cells in an in vitro proliferation assay. Examples of antibodies demonstrating the greatest inhibition activity in this assay as full IgGs are shown in . The antibodies demonstrating the best inhibitory profile as full IgG molecules were hHERmAb-F0178 and hHERmAb-F0179 (); both were demonstrated to specifically recognize HER2 and did not bind to other EGFR family members, including EGFR, HER3, and HER4.
Figure 1. Characterization of anti-HER2 antibodies F0178 and F017, which effectively Inhibits ligand-depended breast cancer cell proliferation. (A) MTS assay examining the effects of 100 nM of control IgG, trastuzumab, pertuzumab, F018, F0179, A0221, E0184 or G0304 on breast cancer cell proliferation in the absence or presence of ErbB ligand HRG. Results are shown as percentage of control cell proliferation. Error bars, SD (B) SDS-PAGE analysis of purified anti-ErbB2 antibodies under non-reducing and reducing conditions. (C) Competitive binding assay. Trastuzumab, pertuzumab, F0178 and F0179 were evaluated for their ability to compete with trastuzumab-FITC or pertuzumab-FITC for binding to BT-474 cells. (D) Amino acid sequences of the insert from antibody-positive phage clones. Sequences were aligned for the consensus motif, which is indicated by underlined letters. (E) Effect of alanine substitutions on antibodies binding to HER2. Data are expressed as means ± SD.
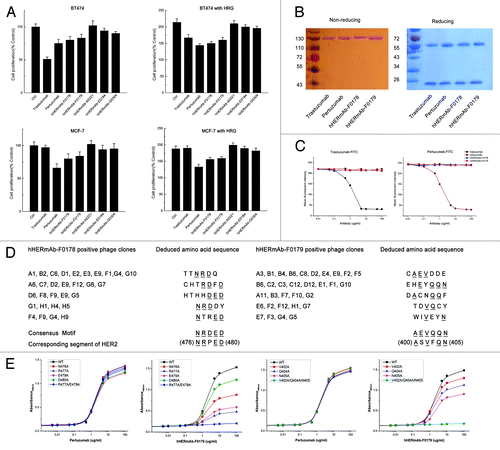
Characterization of anti-ErbB2 antibodies
The molecular weights of the recombinant anti-ErbB2 antibodies hHERmAb-F0178 and hHERmAb-F0179 were determined by SDS-PAGE. Under reducing conditions, each antibody yielded two protein bands with molecular masses of ~55 kDa (heavy chain) and ~25 kDa (light chain). SDS-PAGE analysis under non-reducing conditions showed a single band at ~150 kDa for the F0718 antibody (). The results suggest that these two antibodies are IgG-like molecules, which are composed of two heavy chains and two light chains held together by disulfide bonds. Competitive binding assays were performed and showed that hHERmAb-F0178 and hHERmAb-F0179 have a high binding affinity for ErbB2, and the epitopes of these two antibodies do not appear to overlap with the epitopes of trastuzumab and pertuzumab on ErbB2 (). The affinity constant (Kd) of hHERmAb-F0178 and hHERmAb-F0179 for the ECD of ErbB2 (ErbB2-ECD) was determined by an enzyme-linked immunosorbent assay (ELISA). The data shown in indicate that these two antibodies have an ErbB2 binding affinity comparable to that of trastuzumab and pertuzumab.
Table 1. Binding affinities of recombinant anti-ErbB2 antibodies for ErbB2-ECD
The pharmacokinetics (PK) of trastuzumab, pertuzumab, hHERmAb-F0178 and hHERmAb-F0179 were determined after single-dose intravenous administration to mice. The serum concentrations of these anti-ErbB2 mAbs were measured by ELISA; briefly, we identified the antibodies in mice serum that would bind to ErbB2-ECD immobilized on ELISA plates. As summarized in , the main PK parameters of hHERmAb-F0178 and hHERmAb-F0179 in mice were very close to those of trastuzumab and pertuzumab. The data presented in also showed that hHERmAb-F0178 and hHERmAb-F0179 had PK properties similar to those of a conventional IgG molecule, suggesting that they are highly stable in vivo.
Table 2. Pharmacokinetic parameters of recombinant anti-ErbB2 antibodies in mice
The epitopes of F0178 and F0179 overlap with Fab37
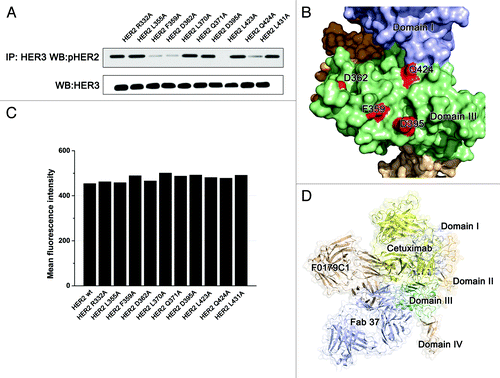
To identify the epitopes recognized by hHERmAb-F0178 and hHERmAb-F0179, phage clones were isolated by panning the Ph.D.-7 phage display peptide libraryCitation16 with hHERmAb-F0178 and hHERmAb-F0179. Three rounds of selection were performed, and, at each round, the library was pre-cleared with a control mouse IgG2a,κ antibody. After the third round of panning, the binding of the isolated phage clones to hHERmAb-F0178 and hHERmAb-F0179 was determined by ELISA. Sequence analysis of hHERmAb-F0178- and hHERmAb-F0179-positive phage clones identified five and six distinct amino acid sequences, respectively (). Alignment of these sequences revealed the consensus motifs NRDED and AEVQQN, which could be aligned with the (476) NRPED (480) and (400) ASVFQN (405) sequence located at ECD III of ErbB2. To determine whether these two sequences within ErbB2 are the epitopes recognized by hHERmAb-F0178 and hHERmAb-F0179, alanine substitutions were introduced into ErbB2-ECD at residues N476, R477, E479, D480, V402, Q404, and N405, and the binding of hHERmAb-F0178 and hHERmAb-F0179 to these ErbB2-ECD mutants was measured by ELISA. The results showed that single alanine substitutions at N476, R477, E479, D480, V402, Q404, or N405 significantly reduced hHERmAb-F0178 and hHERmAb-F0179 binding affinity for ErbB2 (). Double alanine substitutions at positions 477 and 479 further reduced the binding affinity of hHERmAb-F0178, and triple alanine substitutions at positions 402, 404, and 405 almost totally abolished hHERmAb-F0179 binding to ErbB2. In contrast, the binding of pertuzumab to these ErbB2-ECD mutants was approximately the same as its binding to wild-type ErbB2-ECD (). These data demonstrate that the (476) NRPED (480) and (400) ASVFQN (405) sequences within ErbB2 are the hHERmAb-F0178 and hHERmAb-F0179 epitopes.
These epitope mapping and HER2 mutagenesis data revealed that both the hHERmAb-F0178 and hHERmAb-F0179 epitopes were located at ECD III of ErbB2, although they are located a slight distance apart (). These antibody binding sites appeared to be located opposite to the dimerization interfaces in domains II and IV, implying that these antibodies did not directly disrupt the interaction of HER2 with other HERs according to the current HER2 dimerization model.Citation10 Notably, the epitopes of these two antibodies overlap with an anti-domain III antibody, Fab37, described in a previous studyCitation17 (). The effect of these antibodies on HER signaling was further examined.
Figure 2. Optimization of the epitopes and function of anti-domain III antibodies. (A) Epitopes of anti-HER2 antibodies on HER2. Domains I, II, III, and IV of the ErbB2 ECD protomer are colored blue, bright yellow, green, and wheat, respectively. Epitopes of F0178 and F0179 are colored red and yellow, and epitope of Fab37 is colored blue. (B) MCF-7 cells were treated with 100 nM of control IgG or anti-domain III antibodies in the presence of HRG. Co-immunoprecipitation assay and immunoblots assay were assessing ErbB2 signaling and antibodies’ ability to disrupt the formation of ligand-dependent HER2/HER3 heterodimers. (C) Co-immunoprecipitation assay with an ELISA assay to detect pHER2 in MCF-7 cells treated with different scFvs in the in the presence of HRG. (D) MTS assay examining the effects of 100 nM of control IgG, or antibodies on breast cancer cell proliferation in the presence of HRG. Results are shown as percentage of control cell proliferation. Error bars, SD.
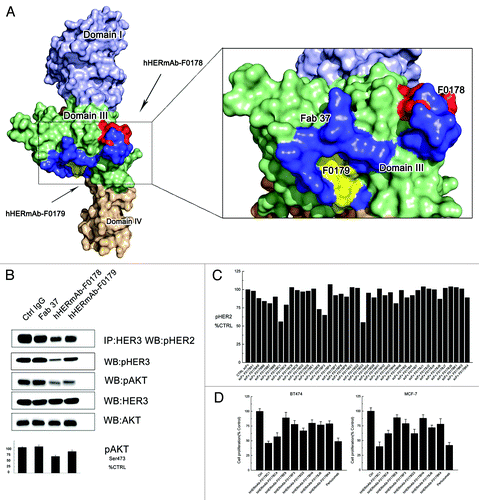
Antibodies with neighboring epitopes vary in their ability to block ErbB2 heterodimerization and signaling
We examined the ability of hHERmAb-F0178, hHERmAb-F0179, and Fab37 to disrupt HRG-dependent ErbB2-HER3 heterodimerization in BT-474 and MCF-7 cell lines. Notably, although these antibodies have similar epitopes, in both cell lines, hHERmAb-F0179 was more effective than hHERmAb-F0178 and Fab37 with respect to disrupting HRG-dependent ErbB2/ErbB3 association and subsequent downstream AKT phosphorylation; however, Fab37 had no detectable effect on the HRG-dependent ErbB2/ErbB3 dimerization (). Although antibody Fc region may contribute this difference, together with the epitope information, a slight epitope shift () to domain III appears to elevate the antibodies’ capacity to disrupt HRG-dependent ErbB2/ErbB3 signaling.
Optimization of the antibodies using a step-by-step epitope refinement method
Because the epitope shift contributes to the inhibition efficacy of the antibody, we employed a step-by-step antibody optimization method to obtain more effective antibodies. Optimization of the scFv corresponding to the lead candidate hHERmAb-F0178 was performed to identify antibodies with enhanced inhibitory activity. Randomized libraries were generated from the hHERmAb-F0178 sequences in which the terminal 6 residues of the VH complementarity-determining region (CDR)2 or CDR3 or the terminal 6 residues of the VL CDR2 or CDR3 were randomly replaced to increase diversity within the VH and VL domains. Optimized scFvs were again selected for binding to human HER2 ECD. The ability of the selected scFv to inhibit the formation of HER2 heterodimerization on the surface of MCF-7 cells was assessed, and scFvs with improved inhibitory profiles were identified (). For full-length IgG1 antibodies have better pharmacokinetic profiles and prospect for targeted therapy, the top 8 scFv candidates were converted to full IgG molecules and assayed for inhibition activity in the BT474 and MCF-7 cell proliferation assay. Most full IgG showed increased activity over the scFv form. The most potent antibody in the inhibition assay, hHERmAbF0178-C1, was selected for further characterization (). The hHERmAbF0178-G3 antibody, which showed the greatest inhibitory activity as a scFv, did not demonstrate the greatest inhibitory activity as a full IgG.
Structure of the ErbB2 extracellular domain in complex with F0178-C1 Fab
Recently, Crystallography study has been employed to investigate precise antibody epitope,Citation18 to further investigate the antibody function and HER2 inhibition mechanism, the crystal structure of the ErbB2 ECD in complex with hHERmAbF0178-C1 Fab was determined using the molecular replacement (MR) method and refined to 3.5 Å resolution with a final Rwork value of 22.1% (Rfree = 25.8%) in the space group P121 ().
Table 3. Data collection and refinement statistics
One HER-Fab complex molecule exists in the asymmetric unit with a Matthews coefficient of 2.8 Å3/Da, corresponding to 56.5% of the solvent content.Citation19 Superimposing HER2 in the HER2-Fab complex in its free form yielded a root–mean–square deviation (r.m.s.d.) of 0.3 Å for all the Cα atoms, indicating that no significant overall structural change occurred, except in several key residues at the antibody–antigen interface ().
Figure 3. The HER2-F0178C1 Fab complex structure and interface. A, ribbon diagrams representations of the F0178C1Fab (left) and the HER2- F0178C1 Fab complex (right) and surface corresponding to the ribbon diagrams shown above with the same color scheme. The light chain and heavy chains of the F0178C1 Fab are colored pink and wheat, respectively, whereas HER2 molecules are colored as . Contact surfaces are highlighted in blue on F0178C1 and red on HER2. B, stereo view of the detailed HER2- F0178C1 Fab interface. The residues that are involved in the intermolecular interaction are shown as colored sticks with the same scheme as the surface representation above; the F0178C1 Fab and HER2 molecules are presented as ribbon diagrams. Dashed lines denote hydrogen bonds.
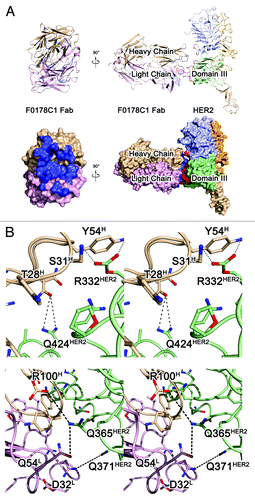
The HER2-Fab complex is approximately 135 Å × 100 Å × 90 Å, and 1219 Å2 of accessible surface area can be observed at the antibody binding interface. The structure of the ErbB2 molecule in the HER2-Fab complex is very similar to its previously reported structure.Citation10,Citation12 The ECD of ErbB2 is composed of four domains, designated I, II, III, and IV (). Domains I and III are structurally very similar: a repeating sequence of hydrophobic residues, mainly leucines, causes these domains to fold into a “β helix” with sides formed by three parallel β sheets. Domains II and IV are also similar in structure, being composed of disulfide-bonded modules (i.e., small structural units held together by one or two disulfide bonds).Citation20 Domain II contains seven disulfide-bonded modules, with an eighth module that is structurally part of domain I. Unlike domain IV, domain II contains an insertion (residues 247–266) in its central disulfide-bonded module. This insertion forms a β hairpin that protrudes from the rest of the protein.Citation10
HERmAbF0178-C1 Fab binds to the pocket formed by domains I and III of HER2, and the functional and structural status of this region has been discussed extensively in previous studies.Citation21,Citation22 The antibody Fab presents a canonical β-sandwich immunoglobulin fold, with the heavy chain folding into the VH and CH domains and the light chain folding into the VL and CL domains. The elbow angle, defined as the subtended angle by two pseudo-2-fold axes relating VH to VL and CH to CL of the antibody Fab, was ~135°. The CDR comprises loops L1, L2, L3, H1, H2, and H3 of HHERmAbF0178-C1, which belong to Chothia canonical classesCitation23 2, 1, 1, 1, 1, and 3, respectively. All the CDR loops of HHERmAbF0178-C1 form a large, deep pocket to accommodate the epitope () and participate in the interaction with HER2.
F0178C1-ErbB2 interactions
The hHERmAbF0178-C1 Fab binds to TNF through a large and highly complementary interface. The epitope of hHERmAbF0178-C1 Fab on HER2 is composed of many discontinuous segments, including residues HER2R12, HER2L13, HER2A15, HER2E330, HER2R332, HER2L355-HER2E357, HER2F359-HER2A364, HER2P369-HER2Q371, HER2A392, HER2D395, HER2S396, and HER2Q424 (). Both the heavy and light chains of hHERmAbF0178-C1 participate in the interaction with HER2, with all the contacts coming from CDRs. Among these CDRs, L3 and H3 contribute to a majority of the interactions with the antigen. Additional contributions are made by CDRs L1, L2, H1, and H2, which indicates a strong and stable interaction within this antigen–antibody pair.
The light chain of hHERmAbF0178-C1 interacts with domain III of HER2. Residue mAbD32 of CDR L3 forms one hydrogen bond with HER2Q371 at domain III of HER2. An extensive network of intermolecular side chain hydrogen bonds between CDR L2 and domain III of HER2 contributes to most of the light chain interactions and positions the side chains of mAbP53, mAbQ54, and mAbP56 of CDR L2 such that they can interact with HER2F359, HER2D395, and HER2S396. CDR L3 additionally contributes to antigen–antibody communication through the hydrogen bond formed between the side chain of mAbW92 and the HER2P363 residue in domain III of HER2 ().
The interface between the heavy chain of hHERmAbF0178-C1 and HER2 is primarily composed of the residues inside domain I and domain III of HER2. The mAbS31 in CDR H1, mAbY54 in CDR H2, and mAbT74 in CDRH3 form hydrogen bonds with HER2R12 and HER2L13 in domain I. Moreover, the residues mAbN52 and AdaD49 of CDR H2 are in contact with HER2D362, HER2D359, and HER2E357 in domain III, as well as HER2A15 in domain I of HER2 ().
The combination of F0178-C1 and pertuzumab more effectively blocks ErbB2/ErbB3 heterodimerization and signaling
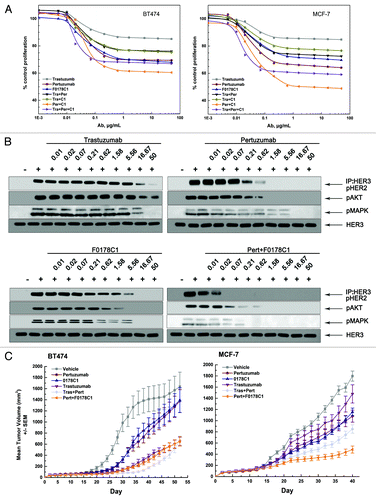
We evaluated the ability of trastuzumab, pertuzumab, hHERmAbF0178-C1, trastuzumab plus pertuzumab, trastuzumab plus hHERmAbF0178-C1, pertuzumab plus hHERmAbF0178-C1, and trastuzumab plus pertuzumab plus hHERmAbF0178-C1 to inhibit the in vitro proliferation of breast cancer cell lines in the presence of HRG. Our data clearly indicated that the antiproliferative activity of these anti-ErbB2 mAbs was directly related to their ability to block ligand dependent ErbB2 heterodimerization and signaling (), as no downregulation of HER2 can be detected with HER2 antibodies (data not shown). Accordingly, pertuzumab plus hHERmAbF0178-C1 demonstrated far more antiproliferative activity than any of the others (). Next, the therapeutic efficacy of anti-ErbB2 mAbs was examined in nude mice bearing established BT474 or MCF-7 xenograft tumors (). Pertuzumab and hHERmAbF0178-C1, but not trastuzumab, significantly delayed MCF-7 tumor progression. Furthermore, the antitumor activity of pertuzumab plus hHERmAbF0178-C1 was similar to that of trastuzumab plus pertuzumab in the BT474 xenograft mouse model. Notably, pertuzumab plus hHERmAbF0178-C1 inhibited tumor growth much more effectively than the combination of trastuzumab and pertuzumab in the MCF-7 xenograft mouse model ().
Figure 4. F0178-C1 effectively Inhibits HER2/HER3-Dependent Signaling and Tumor Growth in Xenograft Models and Shows Increased Inhibition When Combined with Pertuzumab. (A) BT474 and MCF-7 cells were treated with increasing concentration of trastuzumab (gray line), pertuzumab (wine line), F0178-C1 (blue line), the combination of trastuzumab plus pertuzumab (black line), the combination of trastuzumab plus F0178-C1 (dark yellow line), the combination of pertuzumab plus F0178-C1 (orange line), or the combination of trastuzumab plus pertuzumab plus F0178-C1 (violet line) in the presence of HRG (2 nM). Concentrations shown on the x axis reflect the concentration of the single antibody. Cell proliferation relative to untreated control was measured after 4 d using AlamarBlue staining. (B) MCF-7 cells treated with indicated concentrations of trastuzumab, pertuzumab, F0178-C1 or combination of pertuzumab and F0178-C1 were stimulated with 0.5 nM HRG for 10 min. Co-immunoprecipitation assay with an anti-HER3 antibody and blotted for pHER2, and Cell lysates were and immunoblotted to detect pAKT (Ser473), pERK1/2 (Thr202/Tyr204), and total HER3. (C) Mice with established BT474 and MCF-7 cells xenografts were injected intraperitoneally with single dose of pertuzumab (12.5 mg/kg, wine line), trastuzumab (12.5 mg/kg, violet line), F0178-C1 (12.5 mg/kg, blue line), combination of trastuzumab plus pertuzumab (12.5 mg/kg, light gray line)combination of pertuzumab plus F0178-C1 (12.5 mg/kg, orange line), or vehicle (gray line). Data are presented as mean tumor volume ± SEM.
HER2/HER3 heterodimerization and signaling were disrupted by F0178-C1 through a block of HER-ligand binding
It is surprising that anti-domain III antibodies have the capacity to block HER2 heterodimerization because in the current model, domain III did not participate in the dimerization of HER2. However, a structural comparison shows that the epitope of hHERmAbF0178-C1 is similar to that of cetuximab, an anti-EGFR antibody that has been used clinically for a decade. Unlike to other HERs, HER2 was reported to lack of a ligand;Citation24 however, recent studies have suggested that HER2 may have activating membrane-associated ligands.Citation22 Because hHERmAbF0178-C1 bound to domain III and its epitope was similar to that of the ligand blocker, cetuximab, we hypothesized that the antibody-HER2 interaction disrupts the binding of HER2 membrane-associated ligands. Thus, we mutagenesis the residues of HER2 at the antibody-HER2 interface. MCF-7 cells transfected with some HER2 variants with mutations lost the capacity to form phosphorylated HER2/HER3 heterodimers (), without any loss of the surface HER2 level (). These data suggest that HER2 may in fact have a ligand and that hHERmAbF0178-C1 may interrupt this physiological process, which may account for its inhibitory effects.
Figure 5. The F0178C1 epitope may overlap with the HER2-ligand interaction interface. (A) Effects of key residues on the antibody-HER2 interface on HER2 dimerization. (B) Locations of the key residues represents on the right. (C) Fluorescence-activated cell sorting analysis with the 20 ug of FITC labeled antibody pertuzumab. (D) Comparison the relative location of different anti-domain III antibodies. Domains I, II, III, and IV of the ErbB2 ECD protomer are colored as . Antibody F0178C1 is colored wheat and Fab37 is colored bluewhite and modeled use PDB ID 3N85, cetuximab is colored yellow and modeled using PDB ID 1YY
Discussion
Although anti-ErbB2 antibodies have been widely used in breast cancer and gastric cancer therapy, the mechanism underlying the different clinical benefits caused by different epitopes has not yet been elucidated. The inhibition of HER2 dimerization, which blocks the associated signaling, has been reported previously.Citation25 For example, trastuzumab binds to domain IV of HER2,Citation12 and pertuzumab binds to the dimerization arm of HER2.Citation10 The capacity of these antibodies to block the heterodimerization of HER2 is well-understood, and recent studies have carefully evaluated the relationship between the structural epitope and the inhibitory function of the antibody, as domain II and domain IV have their own roles in HER2 dimerization.Citation26 To date, no other HER2-specific antibodies have been reported that have dimerization blocking activity; however, a novel anti-ErbB2 antibody, F0178C1, binds to domain III of HER2, which is far from the dimerization interface in domain II and domain IV of HER2. Regardless, F0178C1 shows excellent heterodimerization blocking activity. These data suggest a unique functional role for domain III, which may be employed as a ligand receiving pocket such as EGFR, HER3, and HER4.
Ligand-induced activation of EGFR involves a marked change in the extracellular region from a ‘tethered’ (inactive) to an ‘extended’ (active) configuration,Citation27 in which an exposed ‘dimerization arm’ in domain II drives the formation of receptor dimers.Citation28 In tethered EGFR, the dimerization arm is occluded by auto-inhibitory intramolecular interactions between domains II and IV, which are also observed in unliganded ErbB3 and ErbB4 but are absent in ErbB2.Citation20 ErbB2 is structurally unique, and even without a bound ligand, its extracellular region resembles the extended (EGF-bound) form of EGFR, with the dimerization arm exposed and apparently ‘poised’ to drive receptor–receptor interaction.Citation28 No known soluble ligand directly regulates ErbB2, and it is the only family member that transforms cells when simply overexpressed (without ligand addition).
Although no exhaustive information on the ligand-independent activation process of HER2 is available, the roles that the regions of HER2 play in this activation process have been reported. For example, recombinant ErbB2 ECD was found to be unable to dimerize in analytical ultracentrifugation studies,Citation29 interactions within the C-terminal region of ErbB receptors are sufficient for constitutive activation,Citation30 and amino acid residues that reside in the kinase domainCitation31 or between the kinase domain and the C-terminusCitation32 are important for dimerization. Taken together, these observations suggest that segments within the intracellular domain contain sequences necessary for dimerization and the ECD of ErbB2 is more likely to be playing the role of dimerization regulation. Although no genuine soluble ligand for ErbB2 is known, at least one membrane-bound regulator that contains EGF-like domains has been identified.Citation33 A subunit of Muc4 (ASGP2) was reported to interact with ErbB2 and to promote its tyrosine phosphorylation. An EGF-like domain in the membrane-associated Muc4 might bind between domains I and III of ErbB2 and induce conformational changes that promote the ability of ErbB2 to form homodimers or heterodimers. Similarly, it was shown in Drosophila that Spitz must be palmitoylated (a modification that drives its membrane association) to regulate dEGFR in vivo.Citation34 Taken together, these data suggest that domain III of HER2 may also play a role as a ligand binding domain. Thus, the inhibition mechanism of anti-domain III antibodies may involve a block of HER2 ligand binding. Confirmation of this mechanism awaits the identification of the ErbB2 ligands. Remarkably, F0178-C1 provides synergistic inhibition of ErbB2 heterodimerization and signaling in combination with pertuzumab, which directly blocks ErbB2 heterodimerization. However, another anti-domain III antibody, Fab 37, did not exhibit the HER2/HER3 dimerization blocking activity,Citation17 partially because the epitope of Fab 37 is slightly farther from domain II. The epitope of cetuximab, an antibody that binds to domain III of EGFR and blocks EGF binding, can illustrate this phenomenon (). Due to the steric hindrance between domains I and III of HER2, ‘cetuximab-like’ anti-HER2 antibodies cannot bind to the area on HER2 as that of cetuximab on EGFR, and F0178C1 may have attained the best relative binding position. Although Herceptin (trastuzumab) is the backbone of HER2-directed breast cancer therapy, recently, more and more data point out that trastuzumab does not block ErbB2 activation but rather disrupts noncanonical ErbB2–ErbB3 interactions that result in constitutive ErbB3 activation,Citation13,Citation35 in this study, we mainly focused on the HRG-dependent ErbB2 activation, and the impact of F0178-C1 on the whole ErbB2 signaling network is ongoing.
In conclusion, our study defines a new class of ErbB2 targeting antibodies that bind to domain III, the probable ErbB2 ligand binding site. Moreover, we employed a step-by-step method to maximize the inhibition effect of these antibodies, which exhibit synergistic inhibition of ErbB2 in combination with pertuzumab, an anti-ErbB2 antibody that direct disrupts heterodimerization. These results offer interesting mechanistic insights into the structure and function of the HER2 receptor. Importantly, F0178C1 demonstrates a unique ability to function synergistically with pertuzumab, suggesting that it has great potential for clinical use.
Materials and Methods
Cell lines, antibodies, and animals
The human breast cancer cell lines BT474 and MCF-7 were obtained from the American Type Culture Collection (ATCC). The cells were cultured in DMEM medium supplemented with 12% fetal calf serum in 5% CO2 at 37.8 °C in a humidified incubator. Trastuzumab was purchased from Roche Ltd and the pertuzumab antibody was expressed and purified using the method described in previous studies.Citation36 Six-week-old female BALB/c mice and five-week-old female BALB/c nude mice were obtained from the Beijing Experimental Animal Center of Chinese Academy of Sciences. All animals were treated in accordance with guidelines of the Committee on Animals of the Chinese Academy of Sciences.
Phage display libraries and selections
Nonimmunized human single-chain variable fragment (scFv) phage display libraries were used for lead-scFv isolation. HER2-ECD, at 10 mg/ml in phosphate buffered saline (PBS), was immobilized on immunotubes (Nunc), and HER2-binding phage was isolated by 3 sequential rounds of panning.Citation37 The optimized variants were isolated by selection from randomized libraries in solution using biotinylated HER2 captured on streptavidin-coated paramagnetic beads (Dynal).Citation38 The F0178 and F0179 VH CDR3 randomized repertoires were constructed by polymerase chain reaction (PCR) using mutagenic oligonucleotides to replace the last 6 VH CDR3 amino acids with randomized codons. The mutated DNA for both lineages was ligated into the phagemid vector pCANTAB6 and electroporated into Escherichia coli TG1.Citation39 Libraries of 6 × 108 and 8 × 108 individual clones were generated for F0178 and F0179, respectively. The randomized libraries were subjected to 1 round of panning on 10 mg/ml of immobilized HER2 ECD, followed by 12 rounds of soluble selection using decreasing concentrations of biotinylated HER2 ECD, from 50 nM down to 100 pM.
Construction, expression and purification
Residues 1–624 of the ErbB2-ECD was prepared as described previouslyCitation40 except that we used the pcDNA3.1(+) expressing vector (Invitrogen) and the FreeStyle 293 expression system (Invitrogen). Antibody screening for a human anti-ErbB2 domain III antibody is discussed below. The heavy and light chains of the antibody was cloned to pcDNA3.1 (+) and expression vectors were co-transfected into Chinese hamster ovary K1 cells using a Lipofectamine 2000 reagent (Invitrogen). The stable transfectants were isolated through limiting dilution in the presence of 600 μg/mL G418 and 300 μg/mL zeocin. The culture supernatants from the individual cell clones were analyzed for antibody production through a sandwich ELISA. The assay used goat anti-human IgG Fc (KPL) as the capture antibodies and goat anti-human kappa-horseradish peroxidase (HRP; Southern Biotechnology Associates) as the detecting antibodies. Purified human IgG1/Kappa (Sigma) was used as the standard control. The clones producing the highest amount of recombinant antibodies were selected and grown in a serum-free medium. The recombinant antibodies were purified through protein A affinity chromatography from the serum-free culture supernatant. The antibody concentrations were determined by absorbance at 280 nm, and the purity was confirmed through SDS–PAGE analysis and western blot.
The Fab fragment of the 0718C1 for the crystallographic investigation was obtained through a papain digestion of an antibody. The digested protein sample was loaded onto a Protein A Sepharose 4 FF column (GE Healthcare). The Fab fragment eluted in the flow-through was separated from the Fc fragment and further purified through ion-exchange chromatography using a Q-Sepharose FF column (GE Healthcare). The protein sample was concentrated to 20 mg/mL and then exchanged to a stock buffer containing 10 mM Tris–HCl (pH 8.0) and 200 mM NaCl. ErbB2-ECD was subsequently mixed with an excess of F0178C1 Fab, and the complex was purified through gel-filtration chromatography (GE Healthcare). This complex was dialyzed against 10 mM Tris–HCl (pH 8.0) and 150 mM NaCl, and was concentrated to 25 mg/ml.
Competitive binding assay
BT474 or MCF-7 cells at 1 × 106 cells/ml were incubated with a subsaturating concentration of the indicated FITC-conjugated anti-HER2 mAbs and increasing concentrations of purified competing antibodies for 1 h at 4 °C. Then, the cells were washed and analyzed by flow cytometry using a FACS can flow cytometer (Becton Dickinson).
Affinity measurement
The affinities of anti-HER2 antibodies for ErbB2-ECD were determined as described previously.Citation41 Briefly, each mAb was incubated with increasing concentrations of ErbB2-ECD for an hour. The concentration of free antibody was then measured by ELISA using immobilized ErbB2-ECD and was used to calculate affinity (Kd).
Pharmacokinetics
Groups of 6-wk-old female BALB/c mice were injected with 5 mg/kg body weight of anti-ErbB2 mAb via the tail vein. Blood samples were taken every day from day 1 to day 15 by retro-orbital bleeding and collected in tubes coated with heparin to prevent clotting. Six mice were used for every time, and each mouse was bled only once. After centrifugation to remove the cells, the plasma samples were stored at –80 °C until analysis. Serum concentrations of anti-ErbB2 mAbs were measured by competitive ELISAs. Briefly, serial dilutions of serum samples were incubated with a subsaturating concentration of trastuzumab-biotin or pertuzumab-biotin on ErbB2-ECD-coated ELISA plates at 37 °C for 1 h. Detection was performed with alkaline phosphatase-conjugated avidin. PK parameters were calculated using a noncompartmental analysis.
Crystallization
Crystallization was performed at 291 K through the hanging-drop vapor-diffusion technique. The crystals were obtained by mixing 1 μl of the protein solution with an equal volume of a reservoir solution. The mixture drop was equilibrated against 500 μl of the reservoir solution. The crystals were obtained with a reservoir solution containing 1.1 M sodium malonate pH 7.0, 0.1 M HEPES 7.0, 0.5% v/v Jeffamine ED-2001, which reached the final dimensions of 100 × 100 × 100 µm3 with the best diffraction within two weeks. The crystals were then cryo-protected through soaking in a cryo-protectant comprising the reservoir solution and 5% glycol. The cryo-protected crystals were subsequently flash-cooled in liquid nitrogen, and then transferred into a dry nitrogen stream at 100 K for X-ray data collection.
X-ray data collection, processing, and structure determination
The diffraction data for the HER2–0178C1 complex was collected at beamline BL5A (Photon Factory) with a resolution of 3.5 Å. The data were processed, integrated, and scaled using the HKL2000 package.Citation42 The crystals belong to space group P1 with cell parameters a = 72.6 Å, b = 82.5 Å, c = 110.3 Å, α = 90°, β = 90°, γ = 90°. The statistics of all data collections and structure refinements are summarized in .
The HER2–0178C1 structure was solved through the molecular replacement method, which employs the crystal structures of HER2 and pertuzumab Fab (PDB code: 1S78) as the initial searching model by using the program PHASER.Citation43 The clear solutions in both the rotation and translation functions indicated the presence of one complex molecule, including one HER2 and one Fab molecule, in one asymmetric unit. This result is consistent with the Matthews coefficient and solvent content.Citation44 The inconsistent residues were manually rebuilt in the program CootCitation45 under the guidance of the Fo–Fc and 2Fo–Fc electron density maps. The residues were refined in PHENIX,Citation46 and the respective working R-factor and R-free decreased from 0.41 and 0.48 to 0.22 and 0.26, respectively, for all data from 50.0 Å to 3.5 Å. The refinement was monitored by calculating Rfree based on a subset containing 5% of the total reflections. Model geometry was verified using the program PROCHECK.Citation47 The data collection and refinement statistics are presented in detail in . All structure figures were prepared using PYMOL.Citation48
HER2 mutagenesis, and antibodies binding
ErbB2 Site-directed mutants (R332A, L355A, F359A, D363A, L370A, Q371A, D395A, L423A, Q424A, L431AA15F, G425F, S429F, N476A, R477A, E479A, D480A, or double mutants R477A/E479A) were created through PCR. All wild-type and ErbB2 mutant constructs used in this study were epitope tagged at the C terminus with a FLAG sequence. All mutations were confirmed by automated DNA sequencing. Two μg/ml of wild-type HER2-WT or HER2-ECD mutants were added to 96-well plates precoated with 5 μg/ml of trastuzumab, followed by incubation at 37 °C for 1 h. The plates were washed, and different concentrations of antibodies or the control antibody pertuzumab were added to each well and incubated at 37 °C for 1 h. After washing, HRP-conjugated goat polyclonal secondary antibody to mouse IgG-H&L was added and the plates were further incubated for 1 h at 37 °C. Finally, TMB was added as a substrate and the absorbance was read at 450 nm.
MCF-7 cells (1 × 106 cells/10 cm dish) were transfected with 1 ug of DNA using Lipofectamine according to the manufacturer’s recommendations (Invitrogen). Twenty-four hours post transfection, cells were serum starved overnight in 0.1% fetal bovine serum/ DMEM at 37 °C prior to treatment.
Coimmunoprecipitation assays
Heterodimer of HER2 was evaluated in a co-immunoprecipitation assay, as previously described.Citation10,Citation11,Citation49 Transfected cells were lysed in 1 ml RPMI lysis buffer (1% v/v DevelopTriton X-100, 1% w/v CHAPS, 10 mM HEPES (pH 7.2), in RPMI medium containing 0.2 mM PMSF, 10 ug/ml leupeptin, 10 U/ml aprotinin, and 1 mM Na3VO4). Homodimers were immunoprecipitated from 500 ul of lysate using mouse anti-V5 antibody(AbD Serotec) covalently coupled to agarose (Pierce ultralink) at 4 °C for 2 h. Complexes were washed twice in lysis buffer and resuspended in SDS sample buffer and boiled. Samples were separated on a 4–12% polyacrylamide gel (Novex) and electro-blotted onto nitrocellulose membranes. Blots were blocked in 10% BSA/TBST and probed with an anti-Flag antibody (F3165, Sigma) to detect HER2 followed by a peroxidase-conjugated anti-mouse secondary antibody (Amersham) To verify and normalize for expression of transfected constructs between experimental conditions, 50 ug of cell lysate was checked by western blotting with different antibodies. Anti-mouse HRP-conjugated secondary antibody was used for visualization by enhanced chemiluminescence (ECL, Amersham Pharmacia Biotech).
Phosphorylation assays
Ligand independent activation of HER2 was detected by a tyrosine phosphorylation assay using an anti-phospho-ErbB2-Tyr1221/1222 antibody (2243; Cell Signaling)
Cell Proliferation Assay
Cells were incubated with different concentrations of recombinant anti-ErbB2 mAbs or peptides for 2 h, followed by the addition of HRG. Recombinant human HRG (R&D Systems) were added at a final concentration of 1 nM, respectively. After an additional 4-d incubation, cell proliferation was determined by Cell Titer 96 AQueous One Solution Cell Proliferation Assay (MTS assay) kit (Promega).
In vivo therapy study
For BT474 or MCF-7 xenograft studies, 3 × 106 BT474 or MCF-7 cells were inoculated into the mammary fat pad of female BALB/c nude mice. When tumor volumes reached an average of about 100 mm3, the mice were randomly divided into groups of 10 mice each. Mice were injected intraperitoneally with single dose of pertuzumab (12.5 mg/kg, wine line), trastuzumab (12.5 mg/kg, violet line), F0178-C1 (12.5 mg/kg mg/kg, blue line), combination of pertuzumab plus F0178-C1 (12.5 mg/kg, orange line), or vehicle (gray line) Tumors were measured with digital calipers, and tumor volumes were calculated by the formula: volume = length × (width)2 /2.
Phage Display Peptide Library Screening
The Ph.D.-7 phage display peptide library (PDPL) kit was purchased from New England BioLabs (Beverley, MA). Biopanning of PDPL with mouse anti-ErbB2 mAbs was performed according to the manufacturer’s instructions. Phage clones were isolated by panning the Ph.D.-7 PDPL with F0178 and F0179. After three rounds of panning, reactivity of positive phage clones with antibodies was measured by ELISA. All antibody positive phage clones were subjected to sequence analysis.
Phage ELISA
ELISA screening of phage clones was performed as previously reported.Citation50 Briefly, 100 μl of supernatant containing amplified particles from each phage clone were added to 96-well plates precoated with mouse anti-ErbB2 mAbs. After incubation for 2 h at RT, detection was performed with HRP-conjugated anti-phage M13 mAb (GE Healthcare). Finally, positive phage clones were subjected to DNA sequence analysis.
Peptide ELISA
Peptides (Control peptide or peptide 355LPESFDG DPASNTAPLQ PE373) were synthesized and conjugated to KLH by Shanghai Science Peptide Biological Technology. Mass Spectrometry and HPLC were employed to confirm the structure and sequence of the peptide. The reactivity of the antibody with peptides was determined by ELISA assay. Briefly, different concentrations of F0178C1 were added to 96-well plates precoated with KLH-conjugated peptide and incubated for 2 h, followed by the addition of HRP-conjugated goat polyclonal secondary antibody to mouse IgG-H&L (Abcam). After washing, 3, 3′, 5, 5′-tetramethylbenzidine (TMB) was added as a substrate and the absorbance
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the staff of Photon Factory, Japan Synchrotron Radiation Facility, and Beijing Synchrotron Radiation Facility for their generous help in collecting X-ray data and Ms. Min Ding, Dr. Yumin Chen, and Dr. Melissa Cruz of Institute of Shanghai Springermedia, for the critical review of the manuscript.
Funding
This work was supported by the Chinese National Natural Science Foundation (Grant Nos.: 81273311, 81172119) and by an AfterTumor Technology Grant (AfterTumor.com Grant No: CN12895NV).
Accession Codes
The coordinates and structural factors of HER2–Fab were deposited in the Protein Data Bank with the accession code 3WSQ.
References
- Hudis CA. Current status and future directions in breast cancer therapy. Clin Breast Cancer 2003; 4:Suppl 2 S70 - 5; http://dx.doi.org/10.3816/CBC.2003.s.018; PMID: 14667277
- Révillion F, Bonneterre J, Peyrat JP. ERBB2 oncogene in human breast cancer and its clinical significance. Eur J Cancer 1998; 34:791 - 808; http://dx.doi.org/10.1016/S0959-8049(97)10157-5; PMID: 9797688
- Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987; 235:177 - 82; http://dx.doi.org/10.1126/science.3798106; PMID: 3798106
- Esteva FJ, Hortobagyi GN, Sahin AA, Smith TL, Chin DM, Liang SY, Pusztai L, Buzdar AU, Bacus SS. Expression of erbB/HER receptors, heregulin and P38 in primary breast cancer using quantitative immunohistochemistry. Pathol Oncol Res 2001; 7:171 - 7; http://dx.doi.org/10.1007/BF03032345; PMID: 11692142
- Yarden Y. The EGFR family and its ligands in human cancer. signalling mechanisms and therapeutic opportunities. Eur J Cancer 2001; 37:Suppl 4 S3 - 8; http://dx.doi.org/10.1016/S0959-8049(01)00230-1; PMID: 11597398
- Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol 2006; 7:505 - 16; http://dx.doi.org/10.1038/nrm1962; PMID: 16829981
- Linggi B, Carpenter G. ErbB receptors: new insights on mechanisms and biology. Trends Cell Biol 2006; 16:649 - 56; http://dx.doi.org/10.1016/j.tcb.2006.10.008; PMID: 17085050
- Ranson M, Sliwkowski MX. Perspectives on anti-HER monoclonal antibodies. Oncology 2002; 63:Suppl 1 17 - 24; http://dx.doi.org/10.1159/000066203; PMID: 12422051
- Kute T, Lack CM, Willingham M, Bishwokama B, Williams H, Barrett K, Mitchell T, Vaughn JP. Development of Herceptin resistance in breast cancer cells. Cytometry A 2004; 57:86 - 93; http://dx.doi.org/10.1002/cyto.a.10095; PMID: 14750129
- Franklin MC, Carey KD, Vajdos FF, Leahy DJ, de Vos AM, Sliwkowski MX. Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex. Cancer Cell 2004; 5:317 - 28; http://dx.doi.org/10.1016/S1535-6108(04)00083-2; PMID: 15093539
- Agus DB, Akita RW, Fox WD, Lewis GD, Higgins B, Pisacane PI, Lofgren JA, Tindell C, Evans DP, Maiese K, et al. Targeting ligand-activated ErbB2 signaling inhibits breast and prostate tumor growth. Cancer Cell 2002; 2:127 - 37; http://dx.doi.org/10.1016/S1535-6108(02)00097-1; PMID: 12204533
- Cho HS, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW Jr., Leahy DJ. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature 2003; 421:756 - 60; http://dx.doi.org/10.1038/nature01392; PMID: 12610629
- Junttila TT, Akita RW, Parsons K, Fields C, Lewis Phillips GD, Friedman LS, Sampath D, Sliwkowski MX. Ligand-independent HER2/HER3/PI3K complex is disrupted by trastuzumab and is effectively inhibited by the PI3K inhibitor GDC-0941. Cancer Cell 2009; 15:429 - 40; http://dx.doi.org/10.1016/j.ccr.2009.03.020; PMID: 19411071
- de Haard HJ, van Neer N, Reurs A, Hufton SE, Roovers RC, Henderikx P, de Bruïne AP, Arends J-W, Hoogenboom HR. A large non-immunized human Fab fragment phage library that permits rapid isolation and kinetic analysis of high affinity antibodies. J Biol Chem 1999; 274:18218 - 30; http://dx.doi.org/10.1074/jbc.274.26.18218; PMID: 10373423
- Wang YX, Jia PY, Wang YX, Qiao YY, Yang RF, Zhou LJ. [Cloning of human scFv against soman transition state analogues (P6) from a large phage antibody library]. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 2010; 26:250 - 2; PMID: 20230690
- Hu S, Liang S, Guo H, Zhang D, Li H, Wang X, Yang W, Qian W, Hou S, Wang H, et al. Comparison of the inhibition mechanisms of adalimumab and infliximab in treating tumor necrosis factor α-associated diseases from a molecular view. J Biol Chem 2013; 288:27059 - 67; http://dx.doi.org/10.1074/jbc.M113.491530; PMID: 23943614
- Fisher RD, Ultsch M, Lingel A, Schaefer G, Shao L, Birtalan S, Sidhu SS, Eigenbrot C. Structure of the complex between HER2 and an antibody paratope formed by side chains from tryptophan and serine. J Mol Biol 2010; 402:217 - 29; http://dx.doi.org/10.1016/j.jmb.2010.07.027; PMID: 20654626
- Hu S, Liang S, Guo H, Zhang D, Li H, Wang X, Yang W, Qian W, Hou S, Wang H, et al. Comparison of the inhibition mechanisms of adalimumab and infliximab in treating tumor necrosis factor α-associated diseases from a molecular view. J Biol Chem 2013; 288:27059 - 67; http://dx.doi.org/10.1074/jbc.M113.491530; PMID: 23943614
- Stopeck AT, Lipton A, Body J-J, Steger GG, Tonkin K, de Boer RH, Lichinitser M, Fujiwara Y, Yardley DA, Viniegra M, et al. Denosumab compared with zoledronic acid for the treatment of bone metastases in patients with advanced breast cancer: a randomized, double-blind study. J Clin Oncol 2010; 28:5132 - 9; http://dx.doi.org/10.1200/JCO.2010.29.7101; PMID: 21060033
- Burgess AW, Cho HS, Eigenbrot C, Ferguson KM, Garrett TP, Leahy DJ, Lemmon MA, Sliwkowski MX, Ward CW, Yokoyama S. An open-and-shut case? Recent insights into the activation of EGF/ErbB receptors. Mol Cell 2003; 12:541 - 52; http://dx.doi.org/10.1016/S1097-2765(03)00350-2; PMID: 14527402
- Alvarado D, Klein DE, Lemmon MA. Structural basis for negative cooperativity in growth factor binding to an EGF receptor. Cell 2010; 142:568 - 79; http://dx.doi.org/10.1016/j.cell.2010.07.015; PMID: 20723758
- Alvarado D, Klein DE, Lemmon MA. ErbB2 resembles an autoinhibited invertebrate epidermal growth factor receptor. Nature 2009; 461:287 - 91; http://dx.doi.org/10.1038/nature08297; PMID: 19718021
- Al-Lazikani B, Lesk AM, Chothia C. Standard conformations for the canonical structures of immunoglobulins. J Mol Biol 1997; 273:927 - 48; http://dx.doi.org/10.1006/jmbi.1997.1354; PMID: 9367782
- Baselga J, Swain SM. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer 2009; 9:463 - 75; http://dx.doi.org/10.1038/nrc2656; PMID: 19536107
- Bublil EM, Yarden Y. The EGF receptor family: spearheading a merger of signaling and therapeutics. Curr Opin Cell Biol 2007; 19:124 - 34; http://dx.doi.org/10.1016/j.ceb.2007.02.008; PMID: 17314037
- Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol 2001; 2:127 - 37; http://dx.doi.org/10.1038/35052073; PMID: 11252954
- Silva F, Cisternas M, Specks U. TNF-α blocker therapy and solid malignancy risk in ANCA-associated vasculitis. Curr Rheumatol Rep 2012; 14:501 - 8; http://dx.doi.org/10.1007/s11926-012-0290-2; PMID: 22956157
- Garrett TP, McKern NM, Lou M, Elleman TC, Adams TE, Lovrecz GO, Kofler M, Jorissen RN, Nice EC, Burgess AW, et al. The crystal structure of a truncated ErbB2 ectodomain reveals an active conformation, poised to interact with other ErbB receptors. Mol Cell 2003; 11:495 - 505; http://dx.doi.org/10.1016/S1097-2765(03)00048-0; PMID: 12620236
- Horan T, Wen J, Arakawa T, Liu N, Brankow D, Hu S, Ratzkin B, Philo JS. Binding of Neu differentiation factor with the extracellular domain of Her2 and Her3. J Biol Chem 1995; 270:24604 - 8; http://dx.doi.org/10.1074/jbc.270.41.24604; PMID: 7592681
- Segatto O, King CR, Pierce JH, Di Fiore PP, Aaronson SA. Different structural alterations upregulate in vitro tyrosine kinase activity and transforming potency of the erbB-2 gene. Mol Cell Biol 1988; 8:5570 - 4; PMID: 2907606
- Chantry A. The kinase domain and membrane localization determine intracellular interactions between epidermal growth factor receptors. J Biol Chem 1995; 270:3068 - 73; PMID: 7531698
- Penuel E, Akita RW, Sliwkowski MX. Identification of a region within the ErbB2/HER2 intracellular domain that is necessary for ligand-independent association. J Biol Chem 2002; 277:28468 - 73; http://dx.doi.org/10.1074/jbc.M202510200; PMID: 12000754
- Nesbitt A, Fossati G, Bergin M, Stephens P, Stephens S, Foulkes R, Brown D, Robinson M, Bourne T. Mechanism of action of certolizumab pegol (CDP870): in vitro comparison with other anti-tumor necrosis factor alpha agents. Inflamm Bowel Dis 2007; 13:1323 - 32; http://dx.doi.org/10.1002/ibd.20225; PMID: 17636564
- Miura GI, Buglino J, Alvarado D, Lemmon MA, Resh MD, Treisman JE. Palmitoylation of the EGFR ligand Spitz by Rasp increases Spitz activity by restricting its diffusion. Dev Cell 2006; 10:167 - 76; http://dx.doi.org/10.1016/j.devcel.2005.11.017; PMID: 16459296
- Sliwkowski MX. Pari passu dimers of dimers. Proc Natl Acad Sci U S A 2012; 109:13140 - 1; http://dx.doi.org/10.1073/pnas.1209959109; PMID: 22851769
- Li B, Shi S, Qian W, Zhao L, Zhang D, Hou S, Zheng L, Dai J, Zhao J, Wang H, et al. Development of novel tetravalent anti-CD20 antibodies with potent antitumor activity. Cancer Res 2008; 68:2400 - 8; http://dx.doi.org/10.1158/0008-5472.CAN-07-6663; PMID: 18381448
- Vaughan TJ, Williams AJ, Pritchard K, Osbourn JK, Pope AR, Earnshaw JC, McCafferty J, Hodits RA, Wilton J, Johnson KS. Human antibodies with sub-nanomolar affinities isolated from a large non-immunized phage display library. Nat Biotechnol 1996; 14:309 - 14; http://dx.doi.org/10.1038/nbt0396-309; PMID: 9630891
- Hawkins RE, Russell SJ, Winter G. Selection of phage antibodies by binding affinity. Mimicking affinity maturation. J Mol Biol 1992; 226:889 - 96; http://dx.doi.org/10.1016/0022-2836(92)90639-2; PMID: 1507232
- He M, Menges M, Groves MA, Corps E, Liu H, Brüggemann M, Taussig MJ. Selection of a human anti-progesterone antibody fragment from a transgenic mouse library by ARM ribosome display. J Immunol Methods 1999; 231:105 - 17; http://dx.doi.org/10.1016/S0022-1759(99)00144-1; PMID: 10648931
- Hudziak RM, Ullrich A. Cell transformation potential of a HER2 transmembrane domain deletion mutant retained in the endoplasmic reticulum. J Biol Chem 1991; 266:24109 - 15; PMID: 1684181
- Carter P, Presta L, Gorman CM, Ridgway JB, Henner D, Wong WL, Rowland AM, Kotts C, Carver ME, Shepard HM. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc Natl Acad Sci U S A 1992; 89:4285 - 9; http://dx.doi.org/10.1073/pnas.89.10.4285; PMID: 1350088
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter Jr. CW, Sweet RM, eds. Macromolecular Crystallography, part A: Academic Press, 1997:307-26.
- McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr 2005; 61:458 - 64; http://dx.doi.org/10.1107/S0907444905001617; PMID: 15805601
- Matthews BW. Solvent content of protein crystals. J Mol Biol 1968; 33:491 - 7; http://dx.doi.org/10.1016/0022-2836(68)90205-2; PMID: 5700707
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 2004; 60:2126 - 32; http://dx.doi.org/10.1107/S0907444904019158; PMID: 15572765
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 2002; 58:1948 - 54; http://dx.doi.org/10.1107/S0907444902016657; PMID: 12393927
- Laskowski R, MacArthur M, Moss D, Thornton J. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst 1993; 26:283 - 91; http://dx.doi.org/10.1107/S0021889892009944
- DeLano W. The PyMOL Molecular Graphics System 2002
- Muthuswamy SK, Gilman M, Brugge JS. Controlled dimerization of ErbB receptors provides evidence for differential signaling by homo- and heterodimers. Mol Cell Biol 1999; 19:6845 - 57; PMID: 10490623
- Perosa F, Luccarelli G, Prete M, Favoino E, Ferrone S, Dammacco F. Beta 2-microglobulin-free HLA class I heavy chain epitope mimicry by monoclonal antibody HC-10-specific peptide. J Immunol 2003; 171:1918 - 26; PMID: 12902494
