Abstract
N-glycosylation is a complex post-translational modification with potential effects on the efficacy and safety of therapeutic proteins and known influence on the effector function of biopharmaceutical monoclonal antibodies (mAbs). Comprehensive characterization of N-glycosylation is therefore important in biopharmaceutical development. In early development, e.g. during pool or clone selection, however, only minute protein amounts of multiple samples are available for analytics. High sensitivity and high throughput methods are thus needed. An approach based on 96-well plate sample preparation and nanoLC-MS of 2- anthranilic acid or 2-aminobenzoic acid (AA) labeled N-glycans for the characterization of biopharmaceuticals in early development is reported here. With this approach, 192 samples can be processed simultaneously from complex matrices (e.g., cell culture supernatant) to purified 2-AA glycans, which are then analyzed by reversed phase nanoLC-MS. Attomolar sensitivity has been achieved by use of nanoelectrospray ionization, resulting in detailed glycan maps of mAbs and fusion proteins that are exemplarily shown in this work. Reproducibility, robustness and linearity of the approach are demonstrated, making use in a routine manner during pool or clone selection possible. Other potential fields of application, such as glycan biomarker discovery from serum samples, are also presented.
Introduction
N-glycosylation, a complex post-translational modification of proteins, is of central importance in the research and development of therapeutic proteins. Of all approved recombinant biopharmaceuticals, e.g., monoclonal antibodies (mAbs), protein hormones, ~40% are glycoproteins.Citation1 Characterization of N-glycosylation is important during biopharmaceutical process development because N-glycosylation may affect the safety or efficacy of a protein drug.Citation2-Citation6 For mAbs, these effects are based on structural properties derived from the CH2 domain glycosylated at Asn297. Size and charge of attached N-glycans as well as terminal sugar moieties influence complement-dependent cytotoxicity (CDC) and antibody-dependent cell-mediated cytotoxicity (ADCC) potency of IgGs and thereby the overall efficacy. For example, lack of core fucose increases ADCC by improving binding to FcγRIIIa. Increased ADCC activity could be correlated with product safety, i.e., serious infections during TNF-targeted treatment in rheumatoid arthritis patients.Citation7 Moreover, lack of terminal galactose residues and the resulting terminal GlcNAc residues increase CDC by modulating binding to C1q.Citation8 Therefore, it is crucial to analyze the glycan pattern of a biopharmaceutical as early as possible during development to be able to modify the drug candidate, for example by glyco-engineering. Alterations of IgG N-glycosylation have been linked with aging and a variety of diseases, and distinct N-glycans are regarded as potential biomarkers because the interactions of IgGs and Fc-receptors influence and modulate immune responses.Citation9-Citation16
N-glycosylation analysis is sophisticated because of the numerous N-glycan variants that may be attached to the protein molecules and the huge differences in their relative amounts. For example, recombinant human IgG antibodies contain up to 60 different N-glycans with relative amounts of individual N-glycans ranging from 0.02% for an oligomannose structure to more than 70% for the most abundant N-glycan, reflecting differences that cover three orders of magnitude.Citation17 Technologies frequently used for N-glycan analysis are CE, HPAEC-PAD, HPLC, MALDI and ESI-MS and various combinations of these technologies.Citation18 LC-MS is an advantageous combination as LC is able to separate glycan mixtures, after which glycan variants can be individually identified and quantified by online MS. However, for various analytical applications, conventional LC-MS is not sufficiently sensitive, especially for cases where sample amount is strongly limited. During early biopharmaceutical development (e.g., pool or clone selection), only minute amounts of recombinant protein from microtiter plates are usually available for protein and glycan analysis.
N-glycan biomarker discovery in patients or healthy individuals is another scenario where sample amount is typically very limited. In proteomics, similar limitations have been circumvented by reducing the dimensions of the analytical system, for example by use of nanoLC-MS. Literature reports of approaches for N-glycan analysis by use of nanoLC-MS are rare. Several investigations reported feasibility of nanoESI or nanoLC for glycan analysis.Citation19-Citation23 Using a separation-free direct infusion nanoESI approach, Prien et al. quantified 2-Citation13[C6]-AA and 2-Citation12[C6]-AA labeled N-glycans relatively, and demonstrated the usefulness of nanoESI for 2-AA glycan analysis.Citation22 Wuhrer et al. miniaturized HILIC-MS to nanoscale for oligosaccharide analysis, analyzing underivatized N-glycans with femtomolar sensitivity.Citation19 Avoiding glycan derivatization shortens sample preparation, but the benefit of improved MS detection due to the label is lost.Citation19,Citation24 Kalay et al. have used normal-phase nano scale HPLC-MS with online fluorescence to analyze 2-AB N-glycans.Citation20 However, their approach resulted in long and time consuming gradients to achieve a good chromatographic resolution. Ritamo et al. recently published on glycoanalysis utilizing nano-reversed phase chromatography (RPC).Citation21 A nanoLC system was used to separate permethylated N-glycans, achieving separation for various structural isomers on a nano RP-column. Permethylation of N-glycans is an orthogonal approach to labeling. But it requires the use of toxic reagents and also the formation of side products is rather likely which makes routine utilization questionable. Gong et al. used multiplex tandem mass tag labeling for the quantification of neutral IgG N-glycans. They could demonstrate that their approach is suitable for the quantification of major N-glycans and highly reproducibly with linearity over several orders of magnitude.Citation23 However, their short PNGaseF incubation and sample preparation at elevated temperatures might result in incomplete deglycosylation and the loss of sialic acids.Citation25-Citation27 It has been reported previously that RP-LC with online MS is broadly applicable for analysis of differently reducing-end labeled N-glycans.Citation17,Citation28,Citation29 For RP- LC-MS, we have demonstrated that anthranilic acid (2-AA) as label is advantageous for glycan analysis.Citation17 The tag not only improves ionization by contribution of an additional charge carrying residue to the N-glycan, it also improves separation of labeled glycans on RP. The combination of 2-AA labeling with RP separation offers the additional advantage that N-glycans are separated according to their glycan type (high mannose, hybrid and complex), whether they carry a fucose residue at their core N-acteylglucosamine or not and according to their charge caused by terminal sialylation.
Based on these findings we report in this manuscript a new RP nanoLC-MS approach to analyze minute amounts of glycoproteins with high sensitivity. The presented 96-well based sample preparation work-flow is simple, offers high throughput and can be applied during early biopharmaceutical development as a routine or to discover clinical relevant changes of N-glycosylation with high sensitivity. The method has high resolving power and separates many N-glycan structural isomers including multiple branched and acidic variants. In addition, 2-AA labeled glycans can be identified by MSn of the ion-trap mass spectrometer. An overall sensitivity for a single N-glycan on column of ~400 attomol (amol) was observed with the recently introduced MS source. Furthermore, we demonstrate that the method is feasible for N-glycan characterization of minute amounts of mAbs, as well as for N-glycan biomarker discovery. This is shown by comparing the N-glycosylation of an Fc containing therapeutic protein from different cell clones and the method’s broad applicability is demonstrated by glycan analysis of IgGs from human serum. The approach is versatile and due to the use of RPC it should be applicable in many analytical laboratories already working with nanoLC-MS with little effort.
Results
Method development
N-glycosylation analysis of glycoproteins can be performed in different ways by LC-MS: the intact proteins, proteolytic digests or released N-glycans can be analyzed. Released glycans can be derivatized to even the ionization efficiency and thereby to improve the accuracy of quantification. This approach provides excellent coverage of the N-glycosylation pattern. We reported previously a method to identify and quantify 2-AA labeled N-glycans by ion-trap MS after RPC.Citation17 The method is selective for many N-glycan isomers and its robustness and reproducibility have been demonstrated. However, the method was developed to analyze N-glycans of mAbs in advanced development stages, not for early development when limited sample amounts are available and higher sensitivity is required. Thus, we have started to develop a nanoLC-MS method.
Our approach used glycans released from RNase B (Fig. S1 and Table S1), a model protein for N-glycosylation analysis. RNase B N-glycans were prepared as described in the methods section, and the labeled and purified N-glycans were analyzed by nanoLC-MS. The nanoLC was configured in “direct injection on a nano column” mode because highly hydrophilic 2-AA N-glycans like the high mannose type glycans did not bind properly to the trapping column, and were therefore underrepresented in setups that included a pre-concentration step with a trapping column. The user defined injection routine allows injection volumes from 1 to 4 µl without major gradient delay. This is achieved by, depending on the chosen injection volume, switching the sample loop between 5 to 15 min into the flow to ensure that the entire sample leaves the loop before switching it back to loading position. Because of the direct injection, samples must be highly purified to avoid salt plugs entering the nanospray chamber of the MS, which may damage the emitter tip and shorten its lifetime. The chosen acidic mobile phases resulted in high selectivity for many glycan isomers on RP and improved ionization of glycans in the positive ionization mode. The portion of formic acid in the mobile phase could be lowered to 0.5% compared with 1% for the LC-MS method, which may be due to the more efficient ionization in the nano spray. The 2-AA glycans occurred mostly as double [M+2H]2+ charged ions, 2-AA N-glycans smaller than 1500 Da occurred as single [M+H]1+ charged ions. In addition along with protonated ions, mixed adduct ions with sodium (e.g., [M+H+Na]2+) or potassium (e.g., [M+H+K]2+) were also present. The 2-AA glycans were identified by MS, MSCitation2 and MS.Citation3 In addition to RNase B, the method was tested with multiply branched and sialylated glycans to cover all types of glycans. Therefore, several N-glycan standards were labeled with 2-AA and analyzed (Fig. S2). All types of N-glycans can be identified and quantified with this new approach, which is in agreement with our previously reported RP LC-MS approach.Citation17 High mannose glycans elute first, followed by non-fucosylated hybrid and complex variants, then fucosylated hybrid and complex glycans. The overall chromatographic resolution is higher for the nanoLC approach compared with the LC-MS approach.
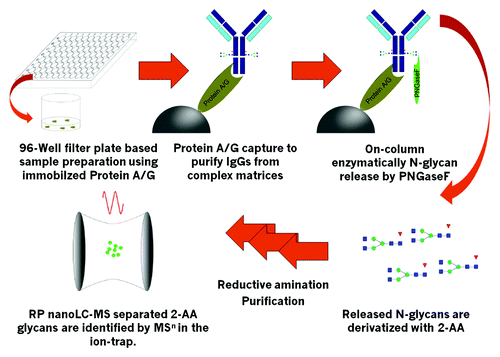
As described in the introduction, the amount of sample available can be very limited at various stages of biopharmaceutical development, especially during early development phases like pool or clone selection. During clone selection, numerous samples in complex matrices (cell culture supernatant) must be analyzed, making affinity purification necessary. Therefore, sample preparation had to be adapted to increase throughput and include affinity purification steps. 96-well plate-based sample preparation is a viable option, the success of which has already been demonstrated for glycoprotein analytics.Citation10,Citation15,Citation30 We selected a centrifugation-based 96-well filter plate sample preparation because we had observed inhomogeneous flow through the small scale columns in the filter plate wells on a vacuum manifold. Protein A Sepharose, which is commercially available and state-of-the-art for downstream processing of Fc-part containing biopharmaceuticals (mAbs and fusion proteins), F was used as affinity resin. The schematic work-flow is illustrated in . Deglycosylation with PNGaseF was performed “on-column.” After washing of bound mAb, N-glycans were released by incubation of the Protein A-mAb complex with PNGaseF at 37 °C followed by elution with H2O, which resulted in higher glycan yields than mAb elution followed by PNGaseF digestion in solution. PNGaseF was subsequently removed by ultrafiltration. Glycans were dried by vacuum centrifugation and 2-AA labeling was performed via reductive amination by use of the non-toxic reductive agent picoline borane.Citation31 Excess label was removed by small scale gel filtration, which was performed in custom-made 96-well plates with Sephadex G-10 resin. The last step is a downscaled procedure based on a previously published purification approach.Citation17,Citation27 This purification is highly efficient because it separates labeled N-glycans from excess 2-AA in a single centrifugation step.
Figure 1. Schematic work-flow. Up to 2x 96 samples can be handled simultaneously. Immobilized Protein A or Protein G is used to capture mAbs, Fc-containing fusion proteins or other IgGs with high specificity. Immobilized target is then highly efficiently deglycosylated with the use of PNGaseF. Released N-Glyans are labeled with 2-AA via reductive amination after ultrafiltration to remove remaining proteins. Labeled and purified 2-AA glycans are identified and quantified by RP nanoLC-MS by use of an ion-trap mass spectrometer.
Qualification of the approach
Several parameters, e.g., sensitivity, robustness, linearity, reproducibility, were investigated to qualify the nanoLC-MS method. Different column batches were tested and the method was executed on different days and by different operators. To determine overall sensitivity and linearity of the method N-glycan standard G0F was labeled with 2-AA as described in the methods section and serial dilutions were prepared to obtain concentrations ranging from ~2.2 pmol/µl to 200 amol/µl. One µl of each dilution was injected. EIC peak areas of 2-AA G0F were used for data interpretation (Fig. S3A). Example MS spectra are shown in Figure S4. EICs of the four smallest amounts that are still in the linear range are shown in magnified view (Fig. S3B). Peak areas were plotted against glycan amount (Fig. S3C and D) and linearity was evaluated by linear regression with R2 = 0.9988, indicating good linear correlation between 2.2 pmol and 800 amol. Injections with 600 and 400 amol 2-AA G0F were still detected, but were not in the linear range. The 200 amol 2-AA G0F injections were not detected anymore. The overall detection limit was thus ~400 amol 2-AA G0F glycan on column, and the lower limit of quantification (LLOQ) was 800 amol. Subsequently, the complete method including sample preparation was validated. Reproducibility and robustness were assessed, for example, by comparing results of two different operators (Table S2). With the determined sensitivity, it is possible to analyze all N-glycans of an IgG using less than 1 µg protein. Varying ionization efficiencies among different N-glycan types was reported previously, especially differences between neutral and sialylated glycans.Citation24 In an earlier work we demonstrated that quantification by fluorescence and MS is identical for neutral N-glycans.Citation17 To further qualify our quantification by MS for acidic N-glycans, we compared the quantitative MS data of three N-glycans, the neutral G2F structure and the single and double sialylated glycans SG2F and S2G2F obtained from a fusion protein with high sialylation to the percentages obtained from the highly sensitive UV-cell in the same nanoLC-MS run (Fig. S5). The SG2F glycan has a lower percentage when determined by MS (MS: 44%; UV: 52%) which might be due to loss of the sialic acid as a small G2F peak is visible under the SG2F peak (Fig. S5A). However, for the double sialylated structure S2G2F, the quantity relative to G2F is identical for MS and UV quantification. These findings show that relative quantification by MS is possible and reliable with the developed method, especially for the intended relative comparison of different pools or clones with the same method.
Glycan mapping of a monoclonal antibody
We then demonstrated the feasibility of the method for characterization of therapeutic glycoproteins. The N-glycosylation pattern of a mAb was analyzed. Sample preparation was successfully performed with less than 1 µg mAb from a drug product formulation. Approximately 1.6 pmol 2-AA labeled N-glycans, corresponding to 800 fmol or 120 ng mAb were finally injected after sample preparation. The resulting EIC is shown in . The most abundant oligosaccharides are complex type N-glycans, G0F followed by the two G1F isomers with 1,3 and 1,6 galactosylation, respectively. Minor abundant species are shown in magnified view (). Identified N-glycans and relative N-glycan composition of the mAb are listed in . The absolute N-glycan amounts range from ~640 amol for a M7 isomer to 1.1 pmol for the most abundant G0F glycan, again showing the huge differences in relative glycan amounts and the resulting requirements to the method in terms of linearity and sensitivity. Listed structures in are deduced from their mass and from MS,Citation2 as well as from MSCitation3 data. For example, the peaks 13, 14 and 15 have the same mass and are not distinguishable by MSCitation2 or MS.Citation3 Peaks 13 and 14 correspond to G1F with the terminal galactose residue on the 1,3 arm or 1,6 arm, respectively, and the separated peak 15, also named G1F, might be a truncated bi-secting or tri-antennary variant with the same number of sugar moieties, but not a third G1F isomer. As described above, the elution order of 2-AA glycans is similar to the pattern obtained by RP LC-MS. More hydrophilic oligomannose structures elute first, followed by non-fucosylated complex variants. Hybrid structures elute before complex type structures as observed for fucosylated glycan structures. Assigned glycan structures are shown in Figure S6.
Figure 2. NanoLC-MS glycan map of a monoclonal antibody. 1.6 pmol of 2-AA labeled N-glycans corresponding to ~120 ng antibody were used for analysis. EICs of the 2-AA labeled N-glycans are depicted. Complete glycan map (A) and magnified view (B) are shown. Peaks are numbered in elution order. Identified glycans are listed in , glycan structures are depicted in Figure S6.
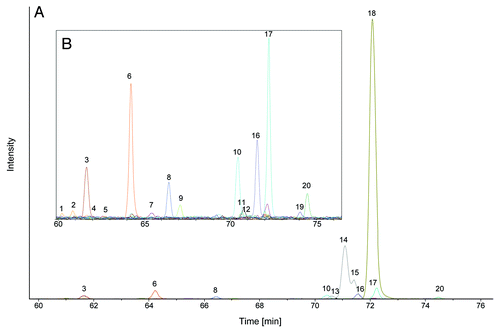
Table 1. 2-AA glycans observed in the glycan map of a monoclonal antibody
Application during early biopharmaceutical development
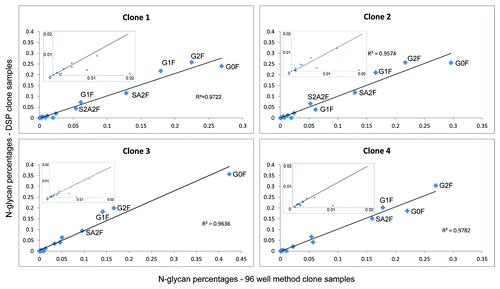
Central goal of our investigation was a glycan characterization approach that can be used in a routine manner during clone selection. In early stages of biopharmaceutical development, protein producing cells (clones) are cultivated in microtiter plates in a few hundred microliters of medium and titers around 1 mg/ml. The aim of this procedure is to identify a clone with appropriate characteristics, for example, distinct protein and glyco-variants and a high titer. Clones must be analyzed comprehensively, but characterization is difficult because as many different aspects of the biopharmaceutical candidate as possible have to be analyzed, hence only minute amounts can be used for each analysis. With the presented work-flow, it is possible to characterize N-glycosylation patterns during clone selection employing only minute amounts of protein. Different clones of an Fc-part containing fusion protein from early development were used to demonstrate the feasibility. Cell culture supernatants from different clones with concentrations between 0.3 mg/ml and 1.7 mg/ml were analyzed. Sample preparation was performed with immobilized Protein A as described in the methods section and shown in . Supernatant (5 µl) from each clone sample containing between 1.5 µg and 8 µg fusion protein, respectively, was mixed with 45 µl PBS before sample preparation. Between 0.27 and 1.5 pmol glycans were injected from each sample, depending on the titer. Detailed glycan maps of four different clones based on EICs are shown exemplarily in . Small differences between the clones can be observed, which demonstrates the need for a sensitive, precise and accurate method to differentiate the clones. Major abundant N-glycans are present in all four clone samples. Some minor abundant N-glycans are absent in some clones and some structures are present only in one clone sample, e.g., SA3F is exclusively present in clone 4. Relative distribution of N-glycan types is also comparable. Clone 1 and clone 2 have 2% oligomannose structures and clone 3 and clone 4 have 1% oligomannose structures. Clone 1 has 47.9% terminal galactosylation and 20.6% terminal sialic acids. Clone 2 has 44.8% terminal galactosylation and 21.4% terminal sialic acids. Clone 3 has slightly less terminal galactosylation and sialylation with 36.9% and 17.5%, respectively. Clone 4 has the highest portion of terminal galactosylation (51.7%) and terminal sialylation (24.1%). These results demonstrate that this robust nanoLC-MS methodology can be used to characterize N-glycans in very early biotechnological development. To further qualify the approach the glycan maps obtained with the newly developed 96-well approach are compared with glycan maps from the same clones after conventional downstream processing (DSP) using Protein A columns. Sample preparation including glycan labeling was performed as described earlier.Citation17 shows the correlation plots of glycan maps. The N-glycan pattern are highly similar to the one obtained using the 96-well-based approach. The plots show the linear correlation of the two methods. These findings further demonstrate the potential of the developed approach in biopharmaceutical development.
Figure 3. Glycan map of four Fc containing therapeutic proteins derived from clone selection phase determined by nanoLC-MS after small scale sample preparation. Clones 1–4 are shown exemplarily. (A) Percentages of the different glycoforms are shown. Error bars indicate variability of the method. Glycosylation pattern of the four clones is similar. (B) Magnified view shows the minor abundant N-glycans
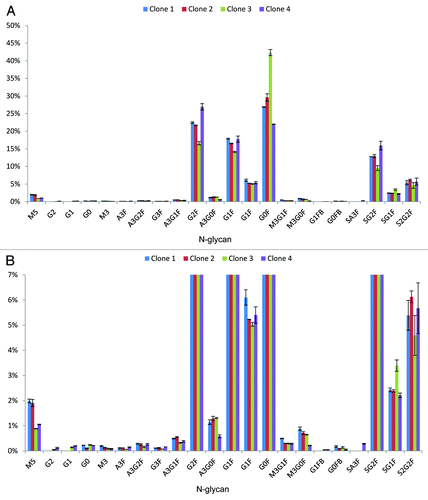
Figure 4. Correlation plots comparing glycan maps of four clones after downstream processing to glycan maps obtained with the newly developed 96-well based nanoLC-MS analysis. Most abundant N-glycans are labeled and linear correlation coefficients are depicted. Insets show the minor abundant N-glycans.
Investigation of serum IgG N-glycans
Application of this new approach is not limited to characterization of biopharmaceutical products and product candidates. Due to its high reproducibility, application in biomarker discovery investigations is possible. Changes in N-glycosylation of glycoproteins such as IgGs have been connected to various diseases and aging in several investigations.Citation13-Citation16 As described in the introduction, terminal galactosylation or fucosylation influence interactions between IgG molecules and Fc-receptors, which in turn affects immune response. This may influence progression, prognosis or outcome of certain diseases. The presented work-flow can be used to screen large populations of patients or healthy subjects for significant differences in their respective N-glycosylation. The high resolving power of the nanoLC-MS method and the order of 2-AA N-glycan elution from the RP column according to their glycan type (e.g., oligomannose, hybrid and complex with and without fucosylation or acidic glycans) allow rapid semi-automatic interpretation of the nanoLC-MS runs. To demonstrate applicability of the method for glycan biomarker discovery, human IgG N-glycosylation was investigated. IgGs were purified from 5 µl of pooled human serum with an estimated IgG concentration of 10 mg/ml with immobilized Protein G Sepharose. In contrast to the above described experiments, the resin was changed from Protein A to Protein G, which allows selective purification of all IgG subclasses 1, 2, 3 and 4. Protein A has no affinity to IgG3 and variable affinity to IgA and IgM, and it is therefore of limited use for this specific study. After Protein G affinity isolation and subsequent sample preparation, a total amount of ~4 pmol 2-AA labeled glycans was analyzed by nanoLC-MS. Resulting chromatograms based on the EICs of the 2-AA N-glycans are shown in . A total of 28 different glycoforms were identified and quantified. Detected glycans, relative glycan composition and the retention time relative to most abundant G0F glycans are listed in ; appropriate glycan structures are shown in Figure S6. The majority of detected N-glycans carry a core fucose, and a small portion carries one to two terminal sialic acids. Complex biantennary glycans carrying a core fucose are the most abundant glycans, comprising more than 70%. Double fucosylated glycan species as G1F2 or G2F2 (Fig. S7) were also found during our investigations and were identified by MSCitation2 by their signature fragments. These findings correspond to previous reports.Citation32
Figure 5. Glycan map of human serum IgGs. (A) EICs of the various 2-AA labeled N-glycans are shown. Peak numbering is according to elution order. lists the identified glycans and their relative amount and Figure S6 shows the glycan structures.
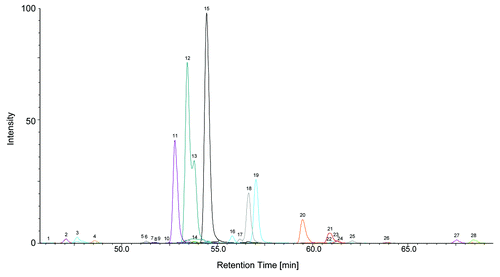
Table 2. 2-AA labeled N-glycans identified from human serum IgGs
With the use of relative retention times and separated 2-AA N-glycans in groups, rapid screening of the N-glycan type composition can be done semi-automatically by integration of the appropriate region of the chromatogram. For example, content of sialic acid-containing and core fucosylated 2-AA glycans can be quantified by integration of peaks between 1.09 and 1.26 relative retention times. With the same methodology, e.g., the degree of fucosylation, which has been associated with certain types of cancer, can be determined.Citation33 The approach is not limited to IgGs. By adjusting sample preparation, other target proteins or protein mixtures can be investigated. These results also show that N-glycan biomarker investigation can be performed with the presented approach.
Discussion
Starting with RNase B (Fig. S1) and several glycan standards (Fig. S2), a new methodology that enables sensitive and selective analysis of less than 1 µg glycoprotein with linearity over several orders of magnitude and high sensitivity for single glycans in the attomolar range has been developed. Compared with a previous publication that reported MS detection of underivatized N-glycans with low femtomolar sensitivity, the presented approach has improved sensitivity, which is most likely due to the applied 2-AA labeling.Citation19 In comparison with the HILIC nanoLC-MS of Kalay et al., a shorter run time with equal or even better chromatographic resolving power was achieved with the RP approach reported here.Citation20
Using our technology, many glycan isomers could be differentiated and different kinds of samples were successfully analyzed with minimal sample consumption. The feasibility of the approach was demonstrated with N-glycan characterization of 160 ng mAb from a drug product formulation with high sensitivity. High sample throughput has been achieved with this 96-well plate sample preparation. A previously published highly efficient gel filtration step has been successfully miniaturized.Citation17,Citation27 This purification step desalts 2-AA glycans efficiently and removes excess label in a single step. Robustness and reproducibility were demonstrated. Requirements for routine use in early biopharmaceutical development are fulfilled, which is shown with the glycan mapping results of clone selection samples. Minor differences in N-glycosylation of a fusion protein from different clones have been detected, which allows early guidance for further development with respect to N-glycosylation. Comparison with glycan maps derived from the same clone samples after conventional downstream processing demonstrated highly similar results.
The nanoLC-MS method has been developed to quantify all types of N-glycans. The formic acid content of the mobile phase was raised to 0.5%. At this concentration sialic acids of N-glycans are mainly protonated and therefore behave like neutral glycans, which enhanced MS intensities in positive ionization mode. Furthermore the used nanoESI source greatly improved and equalized ionization of the labeled N-glycans which was demonstrated by comparing UV to MS data. This ability to identify and quantify all different types of N-glycans, including multiply branched and sialylated variants, facilitates broad application in different areas of glycobiology. One further possible application has been shown exemplarily with the serum IgG glycan biomarker experiment. Results are in good agreement with a previously published investigation by Flynn et al.Citation32
Compared with other recently published high throughput glycan analysis approaches that are also based on 96-well plate sample preparation,Citation34,Citation35 the methodology reported here, which uses 2-AA instead of 2-AB and RP nanoLC-MS instead of UPLC or MALDI MS, results in an up to 10-fold higher sensitivity based on the initial glycoprotein amounts. Higher coverage of the glycan maps is also achieved due to the ability to reliable quantify minor abundant N-glycans. To summarize, the presented method demonstrates the power of RP nanoLC-MS for N-glycosylation analysis. In combination with the high throughput 96-well plate sample preparation procedure N-glycan characterization can be performed in a routine manner during early biopharmaceutical development. The versatility of the method was demonstrated with several different possible fields of application.
Material and Methods
PNGaseF (Roche; 11365177001). Acetonitrile (1.00030.2500) and acetic acid (1.00063.1000) were from Merck. Formic acid (94318), picolineborane, RNase B (R1153) and DMSO (41647) were from Sigma. Protein A Sepharose (17–1297–03), Protein G Sepharose (17–0618–05) and Sephadex® G-10 96-well plats (custom made) were from GE Healthcare. AcroPrep™ Advance Omega™ 10K 96-well filter plates (518–0032) were from Pall. Human serum was from Lonza. MAbs and cell culture supernatants were obtained from in-house development at Sandoz. G0F glycan standard was from Dextra. Complex and acidic N-glycan standards were from TheraProtein.
Purification of IgGs from human serum or cell culture supernatant
Protein G and Protein A Sepharose were used to purify IgG from human pooled serum or cell culture supernatant. A centrifuge with a rotor for 96-well plates (Eppendorf) was used to force liquid through the filter-plates. Serum or cell culture supernatant was applied to a 96-well filter-plate well containing Protein A or Protein G Sepharose, respectively.
Enzymatic N-glycan release by use of PNGaseF
After intensive washing of immobilized IgGs or fusion proteins with PBS N-glycans were enzymatically released with use of PNGaseF and incubation over-night at 37 °C. Released N-glycans were separated from remaining proteins by ultrafiltration using 10K filter plates. Purified N-glycans were brought to dryness in a vacuum centrifuge (Christ RVC 2–25).
Fluorescence labeling of released N-glycans or N-glycan standards
Picolineborane and 2-AA were dissolved in 70% DMSO / 30% acetic acid (v/v) to obtain concentrations of 100 mg/ml and 50 mg/ml, respectively. Labeling solution (15 µl) and deionized H2O (10 µl) were added to either dried N-glycans or lyophilized N-glycan standards. The labeling reaction was performed at 37 °C for 17 h. Excess label was subsequently removed by gel filtration in Sephadex™ G-10 96-well plates. Columns were equilibrated with H2O (800 µl). Samples were filled up to 100 µl with deionized H2O and the sample was applied to the column. After washing with 100 µl of H2O, 2-AA labeled N-glycans were finally eluted with 150 µl of H2O.
NanoLC of labeled N-Glycans
NanoLC (Thermo/Dionex Ultimate 3000) was set-up in “direct injection onto a nano column” mode according to the manufacturer manual. Column compartment with a RP-column (Dionex Acclaim Pepmap 25cm; 75µm I.D.) was held at 40 °C. Mobile phase A consisted of 0.5% formic acid. Mobile Phase B consisted of 0.5% formic acid in 50% ACN. The column was equilibrated with 2% B at a flow rate of 300 nl/min. With a user-defined injection routine, 1–4 µl sample were stacked between loading solution (0.1% formic acid, 1% ACN in ultrapure H2O) in a 20 µl sample loop. Sample loop was switched for 5–15 min, depending on the injected sample volume, in-line to allow the sample to enter the flow, after which the sample loop was switched back to load position to avoid additional gradient delay. Prior to the next injection sample, the loop was washed with loading solution. After sample injection, mobile phase B was raised to 30% over 60 min, then to 95% over 5 min. After holding at 95% B for 5 min, the column was finally re-equilibrated with 2% B for 15 min. Eluting 2-AA N-glycans were detected at 254nm using the variable wavelength detector (Dionex Ultimate 3000 VWD-3400RS with a 3 nl flow cell).
Mass Spectrometry
The outlet of the nanoLC was directly coupled to an ion trap ESI-MS (Bruker AmaZon) equipped with a recently marketed online nano source (Bruker CaptiveSpray NanoBooster™). The ion trap was operated in Enhanced Resolution Mode with a capillary voltage of 1.7 kV. Source temperature was set to 200 °C and a dry gas flow of 3 l/min was used to heat the nano source. MSCitation2 and MSCitation3 spectra were generated with the Auto MSCitation2 mode, Auto MSCitation3 mode and Collision Induced Dissociation (CID). Ion charge control was set to a target value of 2x105 and a maximal accumulation time of 200 ms.
| Abbreviations: | ||
| 2-AA | = | Anthranilic acid or 2-Aminobenzoic acid |
| 2-AB | = | 2-Aminobenzamide |
| ACN | = | Acetonitrile |
| ADCC | = | Antibody-dependent cell-mediated cytotoxicity |
| CE | = | Capillary electrophoresis |
| CDC | = | Complement-dependent cytotoxicity |
| CID | = | Collision induced dissociation |
| EIC | = | Extracted ion chromatogram |
| ESI | = | Electrospray Ionization |
| GlcNAc | = | N-acetylglucosamine |
| HILIC | = | Hydrophilic liquid interaction chromatography |
| HPAEC-PAD | = | High-Performance Anion-Exchange Chromatography with Pulsed Amperometric Detection |
| HPLC | = | High Performance Liquid Chromatography |
| IgG | = | Immunoglobulin G |
| LC-MS | = | Liquid chromatography-Mass spectrometry |
| LLOQ | = | Lower Limit of Quantification |
| mAb | = | Monoclonal antibody |
| MALDI | = | Matrix Assisted Laser Desorption Ionization |
| MS | = | Mass spectrometry |
| PGC | = | Porous graphitized carbon |
| PNGaseF | = | Peptide -N-Glycosidase F |
| RNase B | = | Ribonuclease B |
| RP | = | Reversed phase |
| RPC | = | Reversed phase chromatography |
Additional material
Download Zip (1.3 MB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank Alexander Rysin for technical assistance and Tilen Praper from Analytical Development/Lek Menges for providing the clone samples.
References
- Walsh G. Biopharmaceutical benchmarks 2010. Nat Biotechnol 2010; 28:917 - 24; http://dx.doi.org/10.1038/nbt0910-917; PMID: 20829826
- Chung CH, Mirakhur B, Chan E, Le QT, Berlin J, Morse M, Murphy BA, Satinover SM, Hosen J, Mauro D, et al. Cetuximab-induced anaphylaxis and IgE specific for galactose-alpha-1,3-galactose. N Engl J Med 2008; 358:1109 - 17; http://dx.doi.org/10.1056/NEJMoa074943; PMID: 18337601
- Morell AG, Gregoriadis G, Scheinberg IH, Hickman J, Ashwell G. The role of sialic acid in determining the survival of glycoproteins in the circulation. J Biol Chem 1971; 246:1461 - 7; PMID: 5545089
- Shields RL, Lai J, Keck R, O’Connell LY, Hong K, Meng YG, Weikert SH, Presta LG. Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human Fcgamma RIII and antibody-dependent cellular toxicity. J Biol Chem 2002; 277:26733 - 40; http://dx.doi.org/10.1074/jbc.M202069200; PMID: 11986321
- Jefferis R. Glycosylation as a strategy to improve antibody-based therapeutics. Nat Rev Drug Discov 2009; 8:226 - 34; http://dx.doi.org/10.1038/nrd2804; PMID: 19247305
- Liu C, Dong S, Xu X-J, Yin Y, Shriver Z, Capila I, Myette J, Venkataraman G. Assessment of the quality and structural integrity of a complex glycoprotein mixture following extraction from the formulated biopharmaceutical drug product. J Pharm Biomed Anal 2011; 54:27 - 36; http://dx.doi.org/10.1016/j.jpba.2010.07.044; PMID: 20800406
- Favalli EG, Desiati F, Atzeni F, Sarzi-Puttini P, Caporali R, Pallavicini FB, Gorla R, Filippini M, Marchesoni A. Serious infections during anti-TNFalpha treatment in rheumatoid arthritis patients. Autoimmun Rev 2009; 8:266 - 73; http://dx.doi.org/10.1016/j.autrev.2008.11.002; PMID: 19022409
- Raju TS. Terminal sugars of Fc glycans influence antibody effector functions of IgGs. Curr Opin Immunol 2008; 20:471 - 8; http://dx.doi.org/10.1016/j.coi.2008.06.007; PMID: 18606225
- Abd Hamid UM, Royle L, Saldova R, Radcliffe CM, Harvey DJ, Storr SJ, Pardo M, Antrobus R, Chapman CJ, Zitzmann N, et al. A strategy to reveal potential glycan markers from serum glycoproteins associated with breast cancer progression. Glycobiology 2008; 18:1105 - 18; http://dx.doi.org/10.1093/glycob/cwn095; PMID: 18818422
- Ruhaak LR, Huhn C, Waterreus W-J, de Boer AR, Neusüss C, Hokke CH, Deelder AM, Wuhrer M. Hydrophilic interaction chromatography-based high-throughput sample preparation method for N-glycan analysis from total human plasma glycoproteins. Anal Chem 2008; 80:6119 - 26; http://dx.doi.org/10.1021/ac800630x; PMID: 18593198
- Bones J, Mittermayr S, O’Donoghue N, Guttman A, Rudd PM. Ultra performance liquid chromatographic profiling of serum N-glycans for fast and efficient identification of cancer associated alterations in glycosylation. Anal Chem 2010; 82:10208 - 15; http://dx.doi.org/10.1021/ac102860w; PMID: 21073175
- Ruhaak LR, Uh H-W, Beekman M, Hokke CH, Westendorp RGJ, Houwing-Duistermaat J, Wuhrer M, Deelder AM, Slagboom PE. Plasma protein N-glycan profiles are associated with calendar age, familial longevity and health. J Proteome Res 2011; 10:1667 - 74; http://dx.doi.org/10.1021/pr1009959; PMID: 21184610
- Malhotra R, Wormald MR, Rudd PM, Fischer PB, Dwek RA, Sim RB. Glycosylation changes of IgG associated with rheumatoid arthritis can activate complement via the mannose-binding protein. Nat Med 1995; 1:237 - 43; http://dx.doi.org/10.1038/nm0395-237; PMID: 7585040
- Yamada E, Tsukamoto Y, Sasaki R, Yagyu K, Takahashi N. Structural changes of immunoglobulin G oligosaccharides with age in healthy human serum. Glycoconj J 1997; 14:401 - 5; http://dx.doi.org/10.1023/A:1018582930906; PMID: 9147063
- Wuhrer M, Stam JC, van de Geijn FE, Koeleman CA, Verrips CT, Dolhain RJ, Hokke CH, Deelder AM. Glycosylation profiling of immunoglobulin G (IgG) subclasses from human serum. Proteomics 2007; 7:4070 - 81; http://dx.doi.org/10.1002/pmic.200700289; PMID: 17994628
- Kodar K, Stadlmann J, Klaamas K, Sergeyev B, Kurtenkov O. Immunoglobulin G Fc N-glycan profiling in patients with gastric cancer by LC-ESI-MS: relation to tumor progression and survival. Glycoconj J 2012; 29:57 - 66; http://dx.doi.org/10.1007/s10719-011-9364-z; PMID: 22179780
- Higel F, Demelbauer U, Seidl A, Friess W, Sörgel F. Reversed-phase liquid-chromatographic mass spectrometric N-glycan analysis of biopharmaceuticals. Anal Bioanal Chem 2013; 405:2481 - 93; http://dx.doi.org/10.1007/s00216-012-6690-3; PMID: 23371526
- Mariño K, Bones J, Kattla JJ, Rudd PM. A systematic approach to protein glycosylation analysis: a path through the maze. Nat Chem Biol 2010; 6:713 - 23; http://dx.doi.org/10.1038/nchembio.437; PMID: 20852609
- Wuhrer M, Koeleman CA, Deelder AM, Hokke CH. Normal-phase nanoscale liquid chromatography-mass spectrometry of underivatized oligosaccharides at low-femtomole sensitivity. Anal Chem 2004; 76:833 - 8; http://dx.doi.org/10.1021/ac034936c; PMID: 14750882
- Kalay H, Ambrosini M, van Berkel PHC, Parren PWHI, van Kooyk Y, García Vallejo JJ. Online nanoliquid chromatography-mass spectrometry and nanofluorescence detection for high-resolution quantitative N-glycan analysis. Anal Biochem 2012; 423:153 - 62; http://dx.doi.org/10.1016/j.ab.2012.01.015; PMID: 22330744
- Ritamo I, Räbinä J, Natunen S, Valmu L. Nanoscale reversed-phase liquid chromatography-mass spectrometry of permethylated N-glycans. Anal Bioanal Chem 2013; 405:2469 - 80; http://dx.doi.org/10.1007/s00216-012-6680-5; PMID: 23307132
- Prien JM, Prater BD, Qin Q, Cockrill SL. Mass spectrometric-based stable isotopic 2-aminobenzoic acid glycan mapping for rapid glycan screening of biotherapeutics. Anal Chem 2010; 82:1498 - 508; http://dx.doi.org/10.1021/ac902617t; PMID: 20108906
- Gong B, Hoyt E, Lynaugh H, Burnina I, Moore R, Thompson A, Li H. N-glycosylamine-mediated isotope labeling for mass spectrometry-based quantitative analysis of N-linked glycans. Anal Bioanal Chem 2013; 405:5825 - 31; http://dx.doi.org/10.1007/s00216-013-6988-9; PMID: 23670280
- Ruhaak LR, Zauner G, Huhn C, Bruggink C, Deelder AM, Wuhrer M. Glycan labeling strategies and their use in identification and quantification. Anal Bioanal Chem 2010; 397:3457 - 81; http://dx.doi.org/10.1007/s00216-010-3532-z; PMID: 20225063
- Watanabe T, Inoue N, Kutsukake T, Matsuki S, Takeuchi M. Labeling conditions using a 2-aminobenzamide reagent for quantitative analysis of sialo-oligosaccharides. Biol Pharm Bull 2000; 23:269 - 73; http://dx.doi.org/10.1248/bpb.23.269; PMID: 10726877
- Bigge JC, Patel TP, Bruce JA, Goulding PN, Charles SM, Parekh RB. Nonselective and efficient fluorescent labeling of glycans using 2-amino benzamide and anthranilic acid. Anal Biochem 1995; 230:229 - 38; http://dx.doi.org/10.1006/abio.1995.1468; PMID: 7503412
- Melmer M, Stangler T, Schiefermeier M, Brunner W, Toll H, Rupprechter A, Lindner W, Premstaller A. HILIC analysis of fluorescence-labeled N-glycans from recombinant biopharmaceuticals. Anal Bioanal Chem 2010; 398:905 - 14; http://dx.doi.org/10.1007/s00216-010-3988-x; PMID: 20640408
- Chen X, Flynn GC. Analysis of N-glycans from recombinant immunoglobulin G by on-line reversed-phase high-performance liquid chromatography/mass spectrometry. Anal Biochem 2007; 370:147 - 61; http://dx.doi.org/10.1016/j.ab.2007.08.012; PMID: 17880905
- Prater BD, Connelly HM, Qin Q, Cockrill SL. High-throughput immunoglobulin G N-glycan characterization using rapid resolution reverse-phase chromatography tandem mass spectrometry. Anal Biochem 2009; 385:69 - 79; http://dx.doi.org/10.1016/j.ab.2008.10.023; PMID: 19000897
- Reusch D, Haberger M, Selman MHJ, Bulau P, Deelder AM, Wuhrer M, Engler N. High-throughput work flow for IgG Fc-glycosylation analysis of biotechnological samples. Anal Biochem 2013; 432:82 - 9; http://dx.doi.org/10.1016/j.ab.2012.09.032; PMID: 23026777
- Ruhaak LR, Steenvoorden E, Koeleman CA, Deelder AM, Wuhrer M. 2-picoline-borane: a non-toxic reducing agent for oligosaccharide labeling by reductive amination. Proteomics 2010; 10:2330 - 6; http://dx.doi.org/10.1002/pmic.200900804; PMID: 20391534
- Flynn GC, Chen X, Liu YD, Shah B, Zhang Z. Naturally occurring glycan forms of human immunoglobulins G1 and G2. Mol Immunol 2010; 47:2074 - 82; http://dx.doi.org/10.1016/j.molimm.2010.04.006; PMID: 20444501
- Miyoshi E, Moriwaki K, Nakagawa T. Biological function of fucosylation in cancer biology. J Biochem 2008; 143:725 - 9; http://dx.doi.org/10.1093/jb/mvn011; PMID: 18218651
- Stöckmann H, Adamczyk B, Hayes J, Rudd PM. Automated, high-throughput IgG-antibody glycoprofiling platform. Anal Chem 2013; 85:8841 - 9; http://dx.doi.org/10.1021/ac402068r; PMID: 23919734
- Burnina I, Hoyt E, Lynaugh H, Li H, Gong B. A cost-effective plate-based sample preparation for antibody N-glycan analysis. J Chromatogr A 2013; 1307:201 - 6; http://dx.doi.org/10.1016/j.chroma.2013.07.104; PMID: 23932029
