Abstract
Myeloid-derived suppressor cells (MDSCs), identified as Gr1+CD11b+ cells in mice, expand during cancer and promote tumor growth, recurrence and burden. However, little is known about their role in bone metastases. We hypothesized that MDSCs may contribute to tumor-induced bone disease, and inoculated breast cancer cells into the left cardiac ventricle of nude mice. Disease progression was monitored weekly by X-ray and fluorescence imaging and MDSCs expansion by fluorescence-activated cell sorting. To explore the contribution of MDSCs to bone metastasis, we co-injected mice with tumor cells or PBS into the left cardiac ventricle and Gr1+CD11b+ cells isolated from healthy or tumor-bearing mice into the left tibia. MDSCs didn’t induce bone resorption in normal mice, but increased resorption and tumor burden significantly in tumor-bearing mice. In vitro experiments showed that Gr1+CD11b+ cells isolated from normal and tumor-bearing mice differentiate into osteoclasts when cultured with RANK ligand and macrophage colony-stimulating factor, and that MDSCs from tumor-bearing mice upregulate parathyroid hormone-related protein (PTHrP) mRNA levels in cancer cells. PTHrP upregulation is likely due to the 2-fold increase in transforming growth factor β expression that we observed in MDSCs isolated from tumor-bearing mice. Importantly, using MDSCs isolated from GFP-expressing animals, we found that MDSCs differentiate into osteoclast-like cells in tumor-bearing mice as evidenced by the presence of GFP+TRAP+ cells. These results demonstrate that MDSCs expand in breast cancer bone metastases and induce bone destruction. Furthermore, our data strongly suggest that MDSCs are able to differentiate into osteoclasts in vivo and that this is stimulated in the presence of tumors.
Introduction
Breast cancer, which is the second leading cause of death among women, represents 30% of all malignancies affecting women.Citation1 While much progress has been made in diagnosis and therapeutic options, resulting in improved survival rates, a substantial number of breast cancer patients are still diagnosed with advanced disease, and up to 80% of them develop bone metastases.Citation1,Citation2 Breast cancer bone metastases are primarily osteolytic, inducing intense pain, pathological fractures as well as hypercalemia, and are associated with high mortality rates.Citation3
When tumor cells metastasize to the bone they begin to secrete mediators such as the parathyroid hormone-related protein (PTHrP) that stimulate osteoclast-mediated bone destruction. This results in a release of growth factors including transforming growth factor β (TGFβ) that stimulate the growth of tumor cells and hence the release of additional osteoclast-stimulating factors, in a feed-forward loop.Citation4,Citation5 While many studies have been performed to address the signal transduction cascades that are involved in these late stages of bone destruction, much less is known about the early steps of tumor cell establishment in the bone. Nevertheless, it has become clear that interactions between tumor cells and other cells within the bone microenvironment significantly contribute to the metastatic process, including bone colonization as well as its subsequent destruction.
Myeloid-derived suppressor cells (MDSCs) are a heterogeneous population of myeloid cells at different stages of differentiation that include immature macrophages, granulocytes and dendritic cells as well as myeloid cell precursors at earlier stages of differentiation.Citation6 First dubbed “natural suppressor cells,” MDSCs have a pronounced ability to expand in almost all cancer patients and animal models of malignancy and potently suppress the immune response. They have been the focus of numerous studies in the last few years and now appear as a cell population playing critical roles in promoting multiple aspects of cancer progression, including tumor growth, angiogenesis and metastasis.Citation7 In humans, although MDSCs are commonly defined as LIN–HLA-DR–CD33+ or CD11b+CD14–CD33+ cells, their precise phenotype appears to be much more heterogeneous, varying with cancer types as well as in distinct MDSC subpopulations.Citation8-Citation10
In mice, MDSCs are generally described as Gr1+CD11b+ cells but can be further subdivided into two subpopulations based on the expression of different Gr-1 epitopes, i.e., granulocytic CD11b+Gr-1+ (Ly6G+) and monocytic CD11b+Gr-1+ (Ly6C+Ly6G-) MDSCs. Both these MDSC populations have immunosuppressive activities. In healthy mice, total Gr1+CD11b+ immature myeloid cells (iMCs) represent around 30% and 3% of total bone marrow (BM) and spleen cells, respectively. They are much more abundant in tumor-bearing mice. Indeed, these cells accumulate under the influence of various tumor-derived factors such as the vascular endothelial growth factor (VEGF), TGFβ1, multiple interleukins and prostaglandin E2 (PGE2) and can represent up to 40% of spleenic cells and 80% of BM cells.Citation11-Citation13 Studies focusing on the role of MDSCs in cancer progression showed that the main activity of these cells is to suppress immunity by perturbing both innate and adaptive immune responses,Citation14 identifying them as the main cell population that is responsible for cancer-associated immunosuppression. In fact, it is now well established that MDSCs suppress multiple immune effectors, especially CD4+ and CD8+ T lymphocytes, by inhibiting their proliferation and activation through mechanisms involving arginine I, nitrix oxide metabolism and the overproduction of reactive oxygen species (ROS).Citation15-Citation17 Despite their role in immunosuppression, MDSCs can also directly exert pro-tumorigenic functions by infiltrating into primary lesions and secreting factors such as TGFβ1 or matrix metallopeptidase 9 (MMP9), which have been shown to promote tumor growth, angiogenesis and invasion.Citation18
Based on MDSC cell surface markers, their ability to differentiate into different cell types, and our previous data obtained in models of multiple myeloma,Citation19 we hypothesized that MDSCs could promote the development of breast cancer metastasis to the bone through immunosuppression-independent mechanisms. To test this hypothesis, we chose to employ athymic nude mice, thus eliminating the effects of T cell-MDSC interactions. In this study, we demonstrate that MDSCs play an important role in tumor biology even in the absence of T cells. We also show that, during cancer progression, MDSCs acquire a “tumorigenic” phenotype to promote breast cancer-associated bone resorption in part by differentiating into osteoclasts and also by promoting the expression of osteolytic factors by cancer cells.
Results
MDSCs expand in immunodeficient nude mice
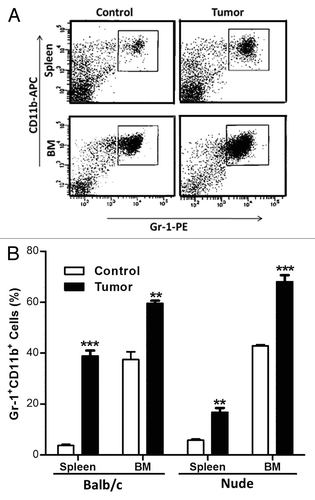
Since MDSC biology is closely related to immunology, we thought it would be critical to first evaluate the use of nude mice as an appropriate model to study MDSCs. To do so, we inoculated immunocompetent BALB/c mice with 4T1 breast cancer cells and immunodeficient nude mice with MDA-MB-231 breast cancer cells. In both models, cancer cells (or PBS as a control) were injected into the 4th mammary fat pad and tumors allowed to grow for approximately 3 weeks. At this point, mice were sacrificed and splenic and BM cells were harvested and labeled with Gr-1-PE and CD11b-APC conjugates for FACS analysis. MDSCs expanded in the spleen and BM of BALB/c mice, representing up to 40% and 60% of the total cells, respectively (), which was in accordance with results previously obtained by others.Citation20 MDSCs also expanded in the spleen and BM of tumor-bearing nude mice. The splenic expansion was more limited in nude mice than in BABL/c mice, Gr1+CD11b+ cells reaching only 20% of total cells. Nevertheless, MDSCs represented up to 68% of total BM cells in tumor-bearing nude mice (), which was comparable with the results obtained in BALB/c mice, giving a first indication that the nude mice model is appropriate to study MDSCs.
Figure 1. Gr-1+CD11b+ cells expand during cancer progression in nude mice as in immunocompetent balb/c mice. Balb/c and nude mice were injected in the 4th mammary fat pad with 4T1 and MDA-MB-231 breast cancer cells respectively or PBS as a control. After 4 weeks of tumor growth, mice were sacrificed. Spleen and BM cells were isolated and Gr-1+CD11b+ cells expansion examined in tumor bearing mice. (A) Spleen and bone marrow of control and tumor nude mice were analyzed by flow cytometry. (B) Quantitative analysis of the expansion of Gr-1+CD11b+ cells in spleens and bone marrow of balb/c and nude mice 4 weeks after mammary fat pad inoculation of 4T1 or MDA-MB-231 breast cancer cells respectively. In both strands, control mice were injected with PBS. Results are presented as the mean ± SEM (5 mice per group)
MDSCs promote cancer growth in immunodeficient nude mice
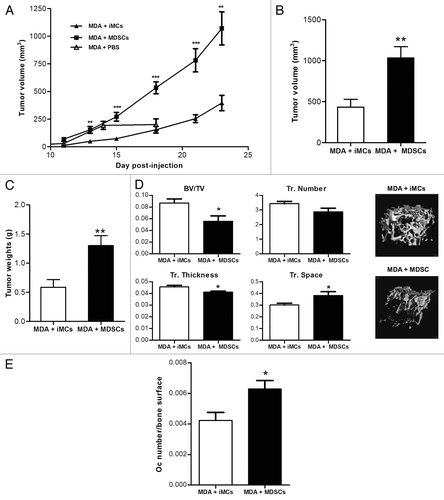
To further investigate the role of MDSCs in tumor progression in nude mice, MDA-MB-231 cancer cells were co-injected in the 4th mammary fat pad of nude mice with 10% (as previously described)Citation20 of either naïve Gr1+CD11b+ iMC sorted from the BM of control mice, or tumor-induced Gr1+CD11b+ MDSCs isolated from tumor-bearing mice, and tumor growth was monitored by means of a common caliper until day 23 post-injection. As observed in , mice co-injected with cancer cells and tumor-induced MDSCs developed neoplastic lesions that reached a final mean volume 2.7-times higher than the final mean volume of lesions developing in mice co-injected with cancer cells and naïve iMCs. These results were confirmed ex vivo by weighing and measuring tumors from both groups (). No significant difference in tumor volume between mice who received cancer cells with PBS or cancer cells with iMCs was observed after 18 d of tumor growth (Fig. S1).
Figure 2. Tumor-induced Gr-1+CD11b+ cells promote tumor cells growth and impair bone mass. One × 106 MDA-MB-231 and 1 × 105 Gr-1+CD11b+ cells isolated from the spleen of control or tumor mice were co-injected in the 4th mammary fat pad of nude mice. Tumor growth was monitored by caliper measurement every 2–3 d (A). At day 24, mice were sacrificed, tumors removed, measured (B) and weighed (C). Results were presented as the mean ± SEM (10 mice per group). (D) Bone mass was assessed by microCT analysis of right femurs from both groups. Data are presented as the mean ± SEM (8 to 10 mice per group).
Upon sacrifice, the hindlimbs and lungs of mice were collected and processed for hematoxylin and eosin (H&E) staining. By microscopic observation of one stained tissue section per mouse, we found that one out of 10 mice that received MDSCs together with cancer cells had lung metastases while no mice co-injected with naïve iMCs did. As a note, bone metastases were observed in neither group (data not shown). Based on these results, it appears that MDSCs do not significantly increase lung metastases from subcutaneous tumors. Nevertheless, it remains possible that metastases were present but not detectable in our examinations. Taken together, our results indicate that MDSCs accumulate and acquire a phenotype that promotes the growth of cancer cells during tumor progression in nude mice. These results were confirmed by observations made in immunocompetent mice, further validating the use of nude mice in our study.
Tumor-induced MDSCs compromises bone mass
Even though no breast cancer cells were detected in the bone, assessment of bone mass by micro-computed X-ray tomography (μCT) analysis showed a significant 30% decrease in the trabecular bone volume of mice injected with the tumor MDSCs compared with the control iMCs. Consistent with reduced bone volume, μCT analyses showed that these mice had less trabeculae, reduced trabecular thickness and increased trabecular spacing (). These data indicate that tumor-induced MDSCs acquire a phenotype that promotes bone loss and that they may create a favorable microenvironment for breast cancer cells to grow and induce osteolysis. Indeed, we observed a higher number of osteoclasts (normalized to the bone surface) in mice that were injected with MDSCs derived from tumor-bearing mice as compared with animals that received control iMCs ().
MDSCs promote breast cancer growth and bone disease under the influence of tumor-derived factors
To test the hypothesis that MDSCs isolated from tumor-bearing mice promote bone metastases and osteolytic lesions more effectively than naïve iMCs, we used the well-characterized model of breast cancer metastasis to the bone in which MDA-MB-231 cells are inoculated into the left cardiac ventricle of athymic nude mice. The mice also received intratibial injections of naïve iMCs or tumor-induced MDSCs previously isolated from the bone marrow of control mice or from mice with mammary fat pad tumors, respectively. All mice were injected with PBS in the contralateral limb. Tumor cell homing to the bone was followed by GFP imaging, and osteolysis was monitored by radiography. As seen in , mice that received MDSCs had significantly larger tumors in the bone and increased osteolytic lesions, after 4 weeks than mice inoculated with naïve iMCs. This indicates that tumor-derived MDSCs are able to promote osteolytic bone destruction. Analysis of TRAP-stained slides as obtained from metastatic bones showed that this effect is due, at least in part, to an increased population of osteoclasts, since mice co-injected with cancer cells and tumor-induced MDSCs showed a significant higher number of osteoclasts (again normalized to the bone surface) when compared with mice receiving cancer cells together with naïve iMCs ().
Figure 3. Tumor-induced Gr-1+CD11b+ cells promote bone metastasis in a model of breast cancer metastasis to bone. One × 105 MDA-MB-231 were inoculated into the left cardiac ventricle in nude mice and 1 × 105 Gr-1+CD11b+ cells isolated from the BM of control or tumor mice were injected in the left tibia of the same mice. The right tibia was injected with PBS as a control. (A) Quantitative analysis of GFP fluorescence detected in mice long bones. (B) Quantitative analysis of osteolytic lesions observed in long bones after radiographs (C) Representative pictures of TRAP staining on bone sections from co-injected mice (left panel) after sacrifice at week 4, and quantitative analysis of the ratio bone surface/osteoclast number (right panel). (D) Osteolytic lesions area were quantified separately in right and left hindlimbs for each mouse.
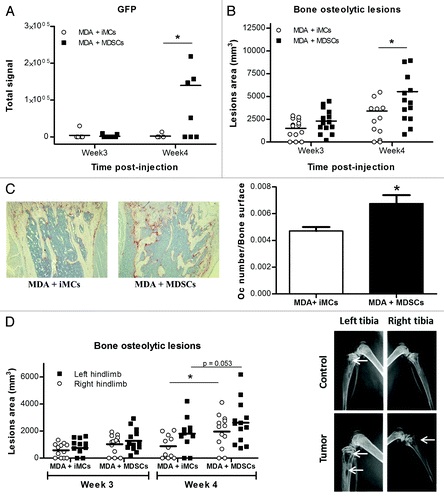
Interestingly, when the areas of osteolytic lesions in both limbs (that injected with GR1+CD11b+ cells and that injected with PBS) were analyzed separately, we observed that (1) both iMCs and MDSCs are able to increase bone resorption locally as compared with PBS, (2) MDSCs induced higher levels of bone resorption than do control iMCs and (3) tumor-derived MDSCs promote bone resorption not only locally but also systemically, since the PBS-injected limbs of MDSCs injected mice developed larger lesions than PBS-injected limbs of the iMCs injected mice ().
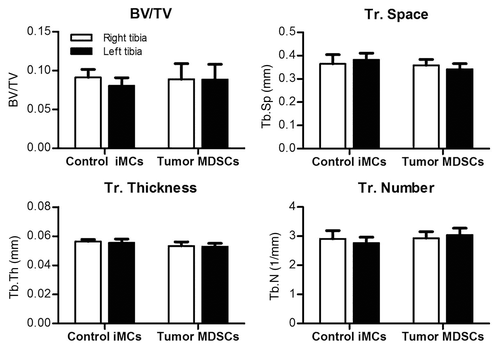
To test whether MDSCs could induce osteolysis in the absence of tumor cells, we performed a similar experiment in which mice were intratibially inoculated only with either naive iMCs or tumor-induced MDSCs. After 3 weeks, mice were sacrificed and bone parameters assessed by μCT. We observed no decrease in overall bone mass in mice who received tumor-induced MDSCs, suggesting that MDSCs can lose their “tumorigenic” phenotype and revert to naïve iMCs when no longer exposed to tumor-derived factors ().
Figure 4. Tumor-induced MDSCs lose their ability to decrease bone mass in the absence of tumor cells. 1 × 105 Gr-1+CD11b+ cells isolated from the BM of control or tumor mice were injected in intratibial in the left tibia. Right tibiaes of the same mice were injected with PBS as a control. Bone parameters were assessed by μCT
Total and monocytic MDSCs expand in the intracardiac model
In order to investigate the mechanisms that could underpin tumor-induced MDSC-promoted osteolysis, we used intracardiac inoculations of nude mice with MDA-MB-231 cancer cells or PBS as a control, and measured MDSC expansion after 2 and 4 weeks. FACS analyses () indicated that, in the intracardiac model, MDSCs expand in the BM, similar to what observed in the MFP model. Splenic MDSC expansion was also observed, but at a lower rate and with no significant difference between the control and tumor-bearing groups. Interestingly, the CD11b+Ly6C+Ly6G- population of monocytic MDSCs expanded to significantly higher extents in tumor mice than in control animals ().
Figure 5. Gr-1+CD11b+ cells expand in a model of breast cancer metastasis to bone and acquire a phenotype that can promote osteolysis. (A) 105 MDA-MB-231 or PBS were injected in IC in nude mice. After 2 and 4 weeks, mice were sacrificed and the Gr-1+CD11b+ cell population in spleen and BM analyzed by flow cytometry. The Gr-1lowCD11b+ population was also analyzed at week4 in BM. (B) Gr-1+CD11b+ were isolated from the BM of control or tumor mice using MACS magnetic microbeads cell sorting and their purity checked. (C-D) Gr-1+CD11b+ cells sorted from control or tumor mice were plated in 6 wells plates and co-cultured with MDA-MB-231 plated in transwells. After 48h, RNA was extracted from both cell types; PTHrP and Gli2 expression in MDA-MB-231 cells (C) and TGF-β expression in Gr-1+CD11b+ cells (D) were analyzed by qRT-PCR.
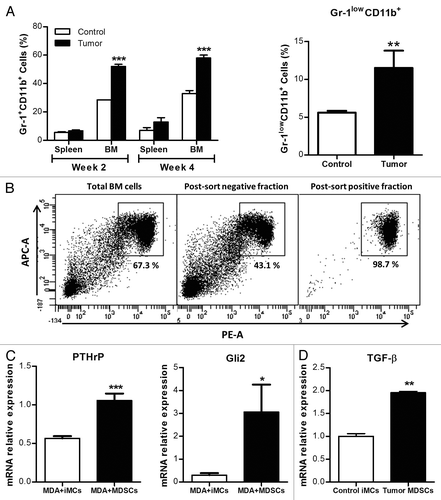
MDSCs express TGFβ1 in BM and induce the expression of osteolytic factors by cancer cells
Gr1+CD11b+ cells were isolated by magnetic cell sorting from the BM of both groups and used for in vitro assessments (). Co-culture experiments of MDA-MB-231 and Gr1+CD11b+ cells followed by quantitative RT-PCR (qPCR), showed that, compared with MDA-MB-231 cells cultured with naïve iMCs, MDA-MB-231 cells cultured with tumor-induced MDSCs express higher levels of GLI2 and PTHRP mRNA, coding for two factors involved in osteoclast activation and in the development of osteolytic lesions (). As TGFβ1 (which stimulates PTHrP expression through GLI2 signaling in MDA-MB-231 cells)Citation21 is known to be secreted by Gr1+CD11b+ cells,Citation18 we assessed its expression (at the mRNA levels) in MDSCs. TGFB1 mRNA expression was indeed 2-fold higher in MDSCs than in naïve iMCs ().
MDSCs can differentiate into active osteoclasts in vitro
Gr1+CD11b+ cells from the BM of both groups isolated as previously () were seeded in 48-well and 96-well plates containing dentine slices. They were then cultured in α-MEM supplemented with 50 ng/mL RANK ligand (RANKL) and 25 ng/mL macrophage colony-stimulating factor (M-CSF). After 12 d of culture, dentine slices were stained with toluidine blue and cells were stained for TRAP expression. We observed that Gr1+CD11b+ cells from both groups were able to differentiate into osteoclasts. However, tumor-induced MDSCs generated more osteoclasts than naïve iMCs, and this result was paralleled by increased dentine resorption ().
Figure 6. Gr1+CD11b+ cells differentiate into osteoclasts in vitro and in vivo. (A) Sorted MDSCs were plated in 48 well plates with dentine discs and cultured in vitro with 25ng/ml of M-CSF and 50ng/ml of RANKL for about 14 d. Wells and dentine discs were then stained and the number of osteoclasts and the resorption area (circled on the zoom panel) quantified. (B) 105 Gr-1+CD11b+ cells isolated from the BM of GFP mice were injected into the left tibia of mice. Mice were co-injected with 106 MDA-MB-231 breast cancer cells or PBS in intracardiac. Four weeks after injections, mice were sacrificed and bones collected for immunostaining with a GFP antibody and for TRAP staining (4 to 8 mice per group). The percentage of total BM positive cells for each staining is represented on the graph (C) Percentage of bone lining cells and TRAP positive cells among GFP+ cells from (B) (left panel). A representative picture of a GFP+ osteoclast is presented on the right panel.
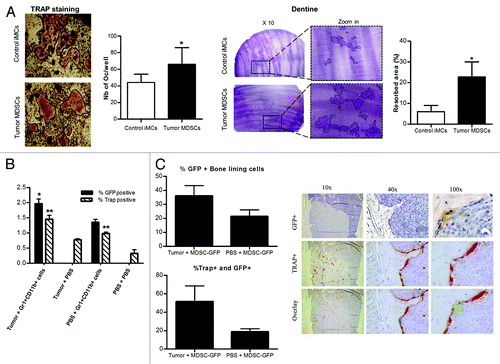
MDSCs differentiate into osteoclasts and increase osteoclast number in vivo
To assess whether MDSCs differentiate into osteoclasts in vivo, Gr1+CD11b+ cells from the BM of GFP-expressing mice were isolated and injected directly into the tibia of nude mice that also received MDA-MB-231 cells or PBS via the intracardiac route. After 4 weeks, mice were sacrificed, and hind limbs were processed for TRAP staining and immunostaining with a GFP-specific antibody. As shown in , we observed a significantly higher percentage of both GFP+TRAP+ cells in tumor-bearing mice compared with control mice. Interestingly, we also observed a higher amount of TRAP+ cells in non-tumor bearing mice that received intratibial injections on Gr1+CD11b+ positive cells as compared with animals receiving PBS, meaning that naïve GR1+CD11b+ cells also increase the number of osteoclasta in the absence of tumor cells. Finally, we observed that in tumor-bearing mice, 37% and 46% of GFP+ cells were multinucleated bone lining cells and TRAP+ cells, respectively, suggesting that MDSCs are able to differentiate into osteoclasts in vivo (). Osteoclast-like GFP+ cells were also detected in the control group but to lower extents (around 20%).
Discussion
As far as we know, our study is the first one showing that MDSCs are able to promote tumor growth in immunodeficient nude mice lacking mature T lymphocytes, the most prominent target for their immunosuppressive functions. Moreover, this is the first study showing that MDSCs can contribute to breast cancer-associated osteolysis. Although it remains unclear whether MDSCs directly attract tumor cells to the bone, it is clear that they locally create a favorable microenvironment for tumor growth.
MDSCs are characterized by their myeloid origin, their immature state and, most importantly, by their ability to suppress immune responses. Indeed, MDSCs have been shown to impair anticancer immune responses in multiple ways, especially by inhibiting cytokine production, T-cell proliferation and by suppressing antigen specific T-cell responses. This latest characteristic is the most studied of the main process involved in the ability of MDSCs to promote cancer growth and metastasis. Nevertheless, we show here that MDSCs promote breast cancer growth and metastasis to bone in nude mice, hence, through a process that is independent from T cells. Indeed, we show that in orthotopic and intracardiac models of breast cancer, MDSCs expand in nude mice and promote tumor growth. MDSCs expansion has previously been shown to occur under the influence of various tumor-derived factors that include—among others—interleukins, VEGF, M-CSF, MMP9 and cyclooxygenase 2.Citation10,Citation13,Citation22-Citation24 Since most of these factors signal through pathways that converge to transcriptional activators of the STAT family, the expansion of MDSCs in nude mice might not be surprising. However, some data suggested that MDSCs enhance tumor angiogenesis and metastasis by modulating the activity of immune cells,Citation18,Citation20 and the ability of MDSCs to promote cancer progression has been mainly linked to their immunosuppressive activity. Our findings that MDSCs promote breast cancer-associated bone osteolysis in immunodeficient mice strongly supports that the role of MDSCs in cancer progression is not limited to their immunosuppressive properties, but also involves their ability to directly interact with and adapt to the tumor microenvironment. Moreover, our results show that immune-independent properties of MDSCs are sufficient to promote breast cancer metastasis to the bone.
In fact, it seems that MDSCs are a source of growth and invasion factors that contribute to tumor growth in the fat pad and bone. They express high level of TGFβ1 in the BM of tumor-bearing mice, inducing the expression of GLI2 and PTHrP by tumor cells (), two factors that are known to promote breast cancer-associated osteolysis.Citation25 Interestingly, Yang et al. have shown that, in primary breast cancer tumors, infiltrating MDSCs express TGFβ1 and promote angiogenesis by secreting MMP9, which in turn enhances VEGF bio-availability (and likely TGFβ1 activation). Thus, MDSCs appear to stimulate cancer cells to express factors that they need to invade and/or to grow at specific sites at least in part through TGFβ1.
We also demonstrate that Gr1+CD11b+ cells are able to differentiate into active osteoclasts, both in vitro and in vivo (). Our data suggest that these osteoclasts are generated by the CD11b+Ly6C+Ly6G- subpopulation of MDSCs. Indeed, this subpopulation is formed by immature monocytic myeloid cells, and it is well established that immediate osteoclasts precursors are cells of the monocyte/macrophage lineage.Citation26 This monocytic subpopulation is less abundant than the granulocytic subpopulation. Nevertheless, it does increase in the intracardiac model of breast cancer and represent around 12% of total BM cells. While this percentage might seem low, an initial increase in bone resorption leads to the release of high amounts of TGFβ in the microenvironment, in turn enhancing PTHrP expression by tumor cells hence initiating a vicious cycle leading to bone destruction. There are additional lines of evidence supporting the idea that MDSCs can differentiate into cell types that tumor cells “need” in their microenvironment. Yang et al. showed that MDSCs recruited into mammary fat pad tumors in mice can differentiate into endothelial cells that are localized in the tumor endothelium of large and small blood vessels.Citation20 Corzo et al. also reported that MDSCs infiltrated in primary tumors rapidly differentiate into tumor-associated macrophages, which contribute to tumor growth.Citation27 Overall, this indicates that MDSCs not only expand during tumorigenesis, but also differentiate or support the differentiation of other cells that contribute to tumor progression. Here, we identified two mechanisms by which MDSCs promote breast cancer bone metastases and associated bone osteolysis. Interestingly, both mechanisms directly or indirectly involve TGFβ overexpression in the bone microenvironment. This suggests that MDSCs participate in the activation and maybe the initiation of a vicious cycle that plays a central role in the development of bone metastases.
Because the orthotopic model of MDA-MB-231 injections in the 4th mammary fat pad doesn’t metastasize to the bone in nude mice, Gr1+CD11b+ cells were mostly studied in the intracardiac model. Nonetheless, we observed that bone mass decreases and that the number of osteoclasts increases in nude mice co-injected with cancer cells and MDSCs as compared with mice co-injected with cancer cells and naïve iMCs, in the mammary fat pad experiment (). This suggests that in the orthotopic model, which best reflects what happens in breast cancer patients in terms of cellular interactions during tumor progression, MDSC expansion causes an increased pool of osteoclast precursors.
It is well accepted that breast cancer cells preferentially metastasize to the bone, because the bone constitutes a favorable soil for their growth and proliferation. Indeed, it has been shown that bone resorption, which releases growth factors such as TGFβ, insulin-like growth factors (IGFs), fibroblast growth factors (FGFs), platelet-derived growth factors (PDGFs) and bone morphogenetic proteins (BMPs), stimulates bone metastasis in several malignancies including breast cancer.Citation28-Citation30 Even though further studies are needed to determine whether the favorable microenvironment created by MDSCs plays a role in the establishment, rather than in the development, of bone metastases, our observation that bone mass is decreased in mammary fat pad tumor-bearing mice co-injected with tumor MDSCs supports this hypothesis. Finally, the finding that tumor-induced MDSCs lose their ability to promote bone destruction in tumor-free mice indicates the tumorigenic phenotype of MDSCs is related to their interactions with cancer cells and is reversible. The reversibility of this process is critical when considering strategies for therapeutic interventions.
Bone metastasis remains an important issue for breast cancer patients, causing high morbidity and mortality. Our work, showing that MDSCs not only exert immunosuppressive functions but also induce bone lesions, identifies this cell population as a critical regulator of breast cancer metastasis to the bone and associated osteolytic bone disease, and as a potential therapeutic target. Recent studies investigating the benefits of targeting MDSCs by inhibiting their expansion or favoring their differentiation into mature cells have already shown promising results.Citation31-Citation33 Further studies are needed to determine the clinical impact of these findings, but our results strongly suggest that targeting MDSCs will limit the negative effects of tumors on the skeleton.
Materials and Methods
Cell lines
Mouse 4T1 cells were purchased from ATCC and MDA-MB-231 cells are a bone-trophic variant developed by Dr. Mundy’s group in San Antonio that has been routinely used by our laboratory and many others.Citation25,Citation34,Citation35 Both cell lines were maintained in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (FBS, Hyclone laboratories) and 1% penicillin/streptomycin (Mediatech). For some experiments, MDA-MB-231 stably transfected with the green fluorescent protein (GFP) obtained as previously described were used.Citation21 They were maintained in the conditions indicated above plus 200 µg/mL G418 (Invitrogen). Cells were cultured in a 37°C atmosphere with 5% CO2.
Mouse models
All procedures were performed with the approval of the Vanderbilt University Institutional Animal Care and Use Committee and in accordance with Federal guidelines.
Mammary fat pad injections
Nude mice were anesthetized using a ketamine/xylazine mixture and positioned ventral side up. Fifty thousand 4T1 cells or 1 × 106 GFP-expressing MDA-MB-231 cells resuspended in 100 µL PBS were then injected into the 4th mammary fat pad of four to five weeks old BALB/c and nude mice, respectively. Control mice were injected with PBS only. Tumor growth was followed by caliper measurement every 2–3 d, and tumor volumes determined using the formula volume = length × (width)2 × 0.5. After 3–4 weeks, mice were sacrificed, tumors removed and spleen and bone marrow cells collected for FACS analysis and Gr-1+CD11b+ cell sorting. For co-injections experiments, MDA-MB-231 cancer cells were injected in nude mice in the same way with 1 × 105 of either naïve or tumor-induced MDSCs.
Intracardiac injections
MDA-MB-231 cells were trypsinized, washed and resuspended in ice-cold PBS at a final concentration of 1 × 106 cells/mL. Nude mice were anesthesized using a ketamine/xylazine mixture and positioned ventral side up. Tumor cells were injected into the left cardiac ventricle using a percutaneous approach with a 27-gauge needle attached to a 1 mL syringe, as described previously. Correct position injection in the left ventricle was confirmed by the appearance of bright red blood at the hub of the needle in a pulsatile fashion. Each mouse received 1 × 105 cells in a 100 µL volume or PBS as a control. Bone metastasis and lesions were followed by GFP fluorescent imaging and X-rays, respectively.
Radiographs of mice
Tumor-bearing animals were sedated using ketamine/xylazine and placed in a prone position. X-ray images were taken at 35kVp for 8 sec using a digital radiography system (Faxitron LX-60). Images were saved and lesions area and numbers were determined using image analysis software (Metamorph, Molecuar Devices, Inc.).
GFP imaging
GFP-labeled tumor cell growth was measured and quantified using the CRi Maestro system. Mice were anesthetized using isoflurane and then placed in the Maestro imaging equipment. After the image was obtained, it was spectrally unmixed to remove the background fluorescence. Images were quantified using region of interest (ROI) analysis software that is supplied with the Maestro system.
Bone histomorphometry
Hind-limb specimens (tibiae and femora) were removed during autopsy and fixed in 10% neutral buffered formalin (Fisher Scientific) for 48 hours at room temperature. Bone specimens were decalcified in 10% EDTA for 2 weeks at 4°C and embedded in paraffin. 5µm-thick sections of bone were stained with hematoxylin & eosin (H+E), orange G, and phloxine for Tumor burden analysis. Sections for osteoclast analysis were stained with Tartrate Resistance Acid Phosphatase (TRAP) (25mg/ml Pararosaniline dye (Sigma) and 0.2mg/ml Naphthol AS-BI substrate (Sigma)). Sections for GFP+ cell analysis were stained with Immunohistochemistry (IHC) (primary antibody: anti-GFP (GeneTex) 1:400 1hr RT, secondary antibody: Goat anti-rabbit IgG-HRP (SantaCruz) 1:400 1hr, RT). Stained femora and tibiae sections were examined under a microscope and quantified using Metamorph software (Molecular Devices, Inc.) and region of interest (ROI) analysis.
Micro-CT analysis
Micro-computed X-ray tomography (µCT) analysis was performed in the Vanderbilt Institute of Small Animl Imaging. The long axis of each specimen was aligned with the scanning axis. One hundred slices from the proximal femur or tibia were scanned at a 12-µm resolution (µCT Scanco Medical, Switerland). The region of interest was trabeculae within the proximal metaphysis of the tibia (0.24 to 1.20 mm) below the growth plate. Images were acquired using 55kV, 114 µA, 300-ms integration and 500 projections per 180° rotation. Contiguous cross sections images of the entire metaphyseal region were acquired. Following reconstruction, the bone tissue was segmented from air soft tissue using a threshold of 270 per thousand, a Gaussian filter of 0.8 and support of 2. Standard architectural characteristics such as trabecular bone volume (BV/TV), trabecular thickness (Tb. Th.), trabecular number (Tb.N.) and t and mean volumetric trabecular space (Tb. Sp.) were calculated using the Scanco evaluation software.
Flow cytometry
BM cells were flushed from the tibia and femur of the mice ad spleens were homogenized in PBS. Cell suspensions were filtered through a 70 μm filter and then incubated for 5 min on ice in red blood cells lysis buffer. After 2 PBS washes, cells were labeled with Gr-1-PE and CD11b-APC fluorescence-conjugated antibodies (Miltenyi Biotec) and analyzed on a flow cytometer BD LSRII (Becton Dickinson). For Gr-1+CD11b+ cell sorting, cells were labeled in the same way and then incubated for 15 min with anti-PE magnetic microbeads. MDSCs were then isolated by running the cell samples on AutoMACS.
In vitro cell differentiation
Ten thousand MDSCs were seeded in 48-well plates and cultured in α-MEM supplemented with 50 ng/mL RANKL and 25 ng/mL M-CSF. Media was changed every 3 d for a 15 d period. At the end of the assay, cells were fixed in ice-cold methanol and stained using a colorimetric TRAcP kit (Sigma-Aldrich) and counter stained in hematoxylin. Multinucleated (more than 3 nuclei) TRAcP cells were counted in each well using a 106 microscopic objective. For osteoclast functionality assays, osteoclasts were grown in the same conditions on dentine discs. At the end of the experiment, dentine discs were removed from culture and sonicated for 2 min in 5 mL 0.25 M ammonium hydroxide to remove cells. The discs were then stained for 5 min 0.05% toluidine blue in 40% MeOH and air-dried. The total resorbed area was calculated using Photoshop image analysis software.
Co-cultures
Ten thousand naïve or tumor-induced MDSCs were seeded in 24-well plates. MDA-MB-231 were seeded in 2 µm wells insert. After 48 h of co-cultures, RNA from MDA-MB-231 and MDSCs was extracted from cells using RNeasy Mini Kit (QIAgen), following manufacturer's instructions.
Quantitative RT-PCR
PTHrP, GLI2 and 18S mRNA expression were measured by quantitative PCR (qPCR). cDNA was synthesized from 1–5 μg of total RNA using SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen) and random hexamers, per manufacturer's instructions. cDNA was serially diluted to create a standard curve and combined with TaqMan Universal PCR Master Mix (Applied Biosystems) and primer: TaqMan PTHLH (Hs00174969_m1), TaqMan GLI2(Hs00257977_m1), or TaqMan Euk 18S rRNA (4352930–0910024; Applied Biosystems). Samples were loaded onto an optically clear 96-well plate (Applied Biosystems) and the qPCR reaction was performed under the following cycling conditions: 50°C for 2 min, 95°C for 10 min, (95°C for 15 sec, 60°C for 1 min) × 40 cycles on a 7300 Real-Time PCR System (Applied Biosystems). qPCR reactions were quantified using the 7300 Real-Time PCR Systems software (Applied Biosystems).
Statistical analyses
All values are expressed as means ± SEM. Values were compared using multifactorial analysis of variance followed by the Student–Newman–Keuls test for multiple comparisons. p < 0.05 was considered significant. * p < 0.05, ** p < 0.01, *** p < 0.005.
| Abbreviations: | ||
| BM | = | bone marrow |
| FACS | = | fluorescence-activated cell sorter |
| iMC | = | immature myeloid cell |
| MDSC | = | myeloid-derived suppressor cell |
| PTHrP | = | parathyroid hormone-related protein |
| TGFβ | = | transforming growth factor β |
Additional material
Download Zip (66 KB)Acknowledgments
The authors would like to thank their mentor and former Director of the Vanderbilt Center for Bone Biology, Dr. Gregory R. Mundy, for his support and input into this project. His training and friendship will never be forgotten. We also thank Dr. Lynn Matrisian and the Vanderbilt University Tumor Micro-Environment Network group for their financial and scientific support, Dr. Daniel Perrien and the Vanderbilt Institute for Imaging Sciences for assistance with the microCT, and Drs. TJ Martin (Melbourne) and Florent Elefteriou (Vanderbilt) for review of this manuscript. We acknowledge the following funding sources: 5P01 CA040035 (G.R.M.), VA Career Development Award (J.A.S.), 5U54 CA 126505 (L.M.M.) and the Philippe Foundation (S.D.).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin 2012; 62:10 - 29; http://dx.doi.org/10.3322/caac.20138; PMID: 22237781
- Yan T, Yin W, Zhou Q, Zhou L, Jiang Y, Du Y, et al. The efficacy of zoledronic acid in breast cancer adjuvant therapy: a meta-analysis of randomised controlled trials. Eur J Cancer 2012; 48:187 - 95; http://dx.doi.org/10.1016/j.ejca.2011.10.021; PMID: 22100904
- Coleman RE. Metastatic bone disease: clinical features, pathophysiology and treatment strategies. Cancer Treat Rev 2001; 27:165 - 76; http://dx.doi.org/10.1053/ctrv.2000.0210; PMID: 11417967
- Yoneda T, Sasaki A, Mundy GR. Osteolytic bone metastasis in breast cancer. Breast Cancer Res Treat 1994; 32:73 - 84; http://dx.doi.org/10.1007/BF00666208; PMID: 7819589
- Sterling JA, Edwards JR, Martin TJ, Mundy GR. Advances in the biology of bone metastasis: how the skeleton affects tumor behavior. Bone 2011; 48:6 - 15; http://dx.doi.org/10.1016/j.bone.2010.07.015; PMID: 20643235
- Nagaraj S, Gabrilovich DI. Myeloid-derived suppressor cells. Adv Exp Med Biol 2007; 601:213 - 23; http://dx.doi.org/10.1007/978-0-387-72005-0_22; PMID: 17713008
- Nagaraj S, Gabrilovich DI. Myeloid-derived suppressor cells in human cancer. Cancer J 2010; 16:348 - 53; http://dx.doi.org/10.1097/PPO.0b013e3181eb3358; PMID: 20693846
- Youn JI, Gabrilovich DI. The biology of myeloid-derived suppressor cells: the blessing and the curse of morphological and functional heterogeneity. Eur J Immunol 2010; 40:2969 - 75; http://dx.doi.org/10.1002/eji.201040895; PMID: 21061430
- Peranzoni E, Zilio S, Marigo I, Dolcetti L, Zanovello P, Mandruzzato S, et al. Myeloid-derived suppressor cell heterogeneity and subset definition. Curr Opin Immunol 2010; 22:238 - 44; http://dx.doi.org/10.1016/j.coi.2010.01.021; PMID: 20171075
- Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol 2009; 9:162 - 74; http://dx.doi.org/10.1038/nri2506; PMID: 19197294
- Bronte V, Apolloni E, Cabrelle A, Ronca R, Serafini P, Zamboni P, et al. Identification of a CD11b(+)/Gr-1(+)/CD31(+) myeloid progenitor capable of activating or suppressing CD8(+) T cells. Blood 2000; 96:3838 - 46; PMID: 11090068
- Melani C, Chiodoni C, Forni G, Colombo MP. Myeloid cell expansion elicited by the progression of spontaneous mammary carcinomas in c-erbB-2 transgenic BALB/c mice suppresses immune reactivity. Blood 2003; 102:2138 - 45; http://dx.doi.org/10.1182/blood-2003-01-0190; PMID: 12750171
- Sinha P, Clements VK, Fulton AM, Ostrand-Rosenberg S. Prostaglandin E2 promotes tumor progression by inducing myeloid-derived suppressor cells. Cancer Res 2007; 67:4507 - 13; http://dx.doi.org/10.1158/0008-5472.CAN-06-4174; PMID: 17483367
- Serafini P, Borrello I, Bronte V. Myeloid suppressor cells in cancer: recruitment, phenotype, properties, and mechanisms of immune suppression. Semin Cancer Biol 2006; 16:53 - 65; http://dx.doi.org/10.1016/j.semcancer.2005.07.005; PMID: 16168663
- Rodriguez PC, Quiceno DG, Zabaleta J, Ortiz B, Zea AH, Piazuelo MB, et al. Arginase I production in the tumor microenvironment by mature myeloid cells inhibits T-cell receptor expression and antigen-specific T-cell responses. Cancer Res 2004; 64:5839 - 49; http://dx.doi.org/10.1158/0008-5472.CAN-04-0465; PMID: 15313928
- Kusmartsev S, Nefedova Y, Yoder D, Gabrilovich DI. Antigen-specific inhibition of CD8+ T cell response by immature myeloid cells in cancer is mediated by reactive oxygen species. J Immunol 2004; 172:989 - 99; PMID: 14707072
- Marigo I, Dolcetti L, Serafini P, Zanovello P, Bronte V. Tumor-induced tolerance and immune suppression by myeloid derived suppressor cells. Immunol Rev 2008; 222:162 - 79; http://dx.doi.org/10.1111/j.1600-065X.2008.00602.x; PMID: 18364001
- Yang L, Huang J, Ren X, Gorska AE, Chytil A, Aakre M, et al. Abrogation of TGF beta signaling in mammary carcinomas recruits Gr-1+CD11b+ myeloid cells that promote metastasis. Cancer Cell 2008; 13:23 - 35; http://dx.doi.org/10.1016/j.ccr.2007.12.004; PMID: 18167337
- Yang L, Edwards CM, Mundy GR. Gr-1+CD11b+ myeloid-derived suppressor cells: formidable partners in tumor metastasis. J Bone Miner Res 2010; 25:1701 - 6; http://dx.doi.org/10.1002/jbmr.154; PMID: 20572008
- Yang L, DeBusk LM, Fukuda K, Fingleton B, Green-Jarvis B, Shyr Y, et al. Expansion of myeloid immune suppressor Gr+CD11b+ cells in tumor-bearing host directly promotes tumor angiogenesis. Cancer Cell 2004; 6:409 - 21; http://dx.doi.org/10.1016/j.ccr.2004.08.031; PMID: 15488763
- Johnson RW, Nguyen MP, Padalecki SS, Grubbs BG, Merkel AR, Oyajobi BO, et al. TGF-beta promotion of Gli2-induced expression of parathyroid hormone-related protein, an important osteolytic factor in bone metastasis, is independent of canonical Hedgehog signaling. Cancer Res 2011; 71:822 - 31; http://dx.doi.org/10.1158/0008-5472.CAN-10-2993; PMID: 21189326
- Melani C, Sangaletti S, Barazzetta FM, Werb Z, Colombo MP. Amino-biphosphonate-mediated MMP-9 inhibition breaks the tumor-bone marrow axis responsible for myeloid-derived suppressor cell expansion and macrophage infiltration in tumor stroma. Cancer Res 2007; 67:11438 - 46; http://dx.doi.org/10.1158/0008-5472.CAN-07-1882; PMID: 18056472
- Talmadge JE, Hood KC, Zobel LC, Shafer LR, Coles M, Toth B. Chemoprevention by cyclooxygenase-2 inhibition reduces immature myeloid suppressor cell expansion. Int Immunopharmacol 2007; 7:140 - 51; http://dx.doi.org/10.1016/j.intimp.2006.09.021; PMID: 17178380
- Kusmartsev S, Gabrilovich DI. Effect of tumor-derived cytokines and growth factors on differentiation and immune suppressive features of myeloid cells in cancer. Cancer Metastasis Rev 2006; 25:323 - 31; http://dx.doi.org/10.1007/s10555-006-9002-6; PMID: 16983515
- Sterling JA, Oyajobi BO, Grubbs B, Padalecki SS, Munoz SA, Gupta A, et al. The hedgehog signaling molecule Gli2 induces parathyroid hormone-related peptide expression and osteolysis in metastatic human breast cancer cells. Cancer Res 2006; 66:7548 - 53; http://dx.doi.org/10.1158/0008-5472.CAN-06-0452; PMID: 16885353
- Burger EH, Van der Meer JW, van de Gevel JS, Gribnau JC, Thesingh GW, van Furth R. In vitro formation of osteoclasts from long-term cultures of bone marrow mononuclear phagocytes. J Exp Med 1982; 156:1604 - 14; http://dx.doi.org/10.1084/jem.156.6.1604; PMID: 7175438
- Corzo CA, Condamine T, Lu L, Cotter MJ, Youn JI, Cheng P, et al. HIF-1α regulates function and differentiation of myeloid-derived suppressor cells in the tumor microenvironment. J Exp Med 2010; 207:2439 - 53; http://dx.doi.org/10.1084/jem.20100587; PMID: 20876310
- Mundy GR. Metastasis to bone: causes, consequences and therapeutic opportunities. Nat Rev Cancer 2002; 2:584 - 93; http://dx.doi.org/10.1038/nrc867; PMID: 12154351
- Percival RC, Urwin GH, Harris S, Yates AJ, Williams JL, Beneton M, et al. Biochemical and histological evidence that carcinoma of the prostate is associated with increased bone resorption. Eur J Surg Oncol 1987; 13:41 - 9; PMID: 3102281
- Suva LJ, Griffin RJ, Makhoul I. Mechanisms of bone metastases of breast cancer. Endocr Relat Cancer 2009; 16:703 - 13; http://dx.doi.org/10.1677/ERC-09-0012; PMID: 19443538
- Lathers DM, Clark JI, Achille NJ, Young MR. Phase 1B study to improve immune responses in head and neck cancer patients using escalating doses of 25-hydroxyvitamin D3. Cancer Immunol Immunother 2004; 53:422 - 30; http://dx.doi.org/10.1007/s00262-003-0459-7; PMID: 14648070
- Kusmartsev S, Cheng F, Yu B, Nefedova Y, Sotomayor E, Lush R, et al. All-trans-retinoic acid eliminates immature myeloid cells from tumor-bearing mice and improves the effect of vaccination. Cancer Res 2003; 63:4441 - 9; PMID: 12907617
- Kusmartsev S, Gabrilovich DI. Role of immature myeloid cells in mechanisms of immune evasion in cancer. Cancer Immunol Immunother 2006; 55:237 - 45; http://dx.doi.org/10.1007/s00262-005-0048-z; PMID: 16047143
- Guise TA, Yin JJ, Taylor SD, Kumagai Y, Dallas M, Boyce BF, et al. Evidence for a causal role of parathyroid hormone-related protein in the pathogenesis of human breast cancer-mediated osteolysis. J Clin Invest 1996; 98:1544 - 9; http://dx.doi.org/10.1172/JCI118947; PMID: 8833902
- Sasaki A, Boyce BF, Story B, Wright KR, Chapman M, Boyce R, et al. Bisphosphonate risedronate reduces metastatic human breast cancer burden in bone in nude mice. Cancer Res 1995; 55:3551 - 7; PMID: 7627963