Abstract
In eukaryotes, ribosome biogenesis is a process of major interest that requires more than 200 factors acting coordinately in time and space. Using genetic and proteomic studies, most of the components have now been identified. Based on its nucleolar localization, we characterized the protein encoded by the open reading frame YGR251W, we renamed Nop19p as playing an essential role in ribosome biogenesis. Depletion of the Nop19p in yeast impairs pre-rRNA processing at sites A0, A1 and A2, leading to a strong decrease in 18S rRNA and 40S subunit levels. Nop19p is a component of 90S preribosomes which assembly is believed to result from stepwise incorporation of UTP modules. We show that Nop19p depletion does not impair the incorporation of UTP subcomplexes on preribosomes and conversely that depletion of UTP subcomplexes does not affect Nop19p recruitment on 90S preribosomes. TAP experiments under stringent conditions revealed that Nop19p interacts preferentially with the DEAH-box RNA helicase Dhr2p and Utp25p, both required for A0, A1 and A2 cleavages. Nop19p appeared essential for the incorporation of Utp25p in preribosomes. In addition, our results suggest that in absence of Nop19p, Dhr2p remains trapped within aberrant preribosomes.
Introduction
The eukaryotic ribosome is a large ribonucleoprotein (RNP) particle that assembles from small (40S) and large (60S) subunits during translation initiation. Ribosome biogenesis is a highly complex process that starts in a specialized nuclear subcompartment, the nucleolus. Late maturation steps take place in the nucleoplasm and in the cytoplasm, following nucleocytoplasmic export (reviewed in ref. Citation1–Citation6). The 40S subunit is assembled around the 18S rRNA, whereas the 60S particle contains the 25S, 5.8S and 5S rRNAs. Three rRNAs (18S, 25S and 5.8S) are processed from a large precursor transcribed by RNA polymerase I, the 35S pre-rRNA (). Pre-rRNA processing involves a series of exonuclease and endonuclease steps (depicted in ) that eliminate external transcribed spacers (5′ETS and 3′ETS) and internal transcribed spacers (ITS1 and ITS2). The 5S rRNA is synthesized independently by RNA Pol III and is incorporated within 90S preribosomes as part of a small ribonucleoprotein particle.Citation7 Moreover, ∼100 nucleotide modifications are introduced in ribosomal RNAs, most of which are catalyzed by the small nucleolar RNPs (snoRNPs) (reviewed in ref. Citation6 and Citation8–Citation10). Concomitantly with these processing events, the ribosomal proteins are progressively incorporated into the maturing particles. In Saccharomyces cerevisiae, more than 180 nonribosomal RNAs and proteins have been shown to participate directly in post-transcriptional steps of ribosome biogenesis. The use of genetic and proteomic analyses allowed the prediction of several distinct RNP intermediates in the pathway of 40S and 60S ribosome subunit synthesis (reviewed in ref. Citation3–Citation6). Among these, the small-subunit (SSU) processome, a 2.2 MDa RNP complex containing the U3 snoRNA and more than 30 proteins (which associate transiently with the preribosomal particles and are not present in the mature ribosomes) is required for the three early pre-rRNA cleavages at sites A0, A1 and A2 (reviewed in ref. Citation6, Citation11–Citation14). The loss of any component of the U3 processome therefore results in a direct cleavage at site A3. The consequence is a loss of the 20S and 18S rRNA concomitantly with an accumulation of the unprocessed 35S and 23S pre-rRNAs (running from the 5′ end of the 35S up to site A3). In parallel, maturation of the 27SA into 5.8S and 25S rRNA remains unaffected. Although many components of the 90S preribosomes have been identified and characterized, it remains unclear how they cotranscriptionally assemble with the nascent transcript to yield a processing-competent particle. Recent evidences suggest that assembly of the 90S preribosomes results from the sequential cotranscriptional association of stable subcomplexes with the nascent 35S pre-rRNA.Citation15 The t-Utps/Utp-A subcomplexCitation15–Citation17 as well as Mrd1pCitation18 are believed to be the earliest factors to associate with the nascent transcript. Indeed, these factors associate co-transcriptionally and their incorporation appears to be a prerequisite for the subsequent recruitment of other subcomplexes. The Pwp2/Utp-B and the Utp-C modulesCitation19–Citation21 are the next two subcomplexes to associate with the nascent transcript. Their order of incorporation still remains unclear as they can assemble independently from one other.Citation15 The association of Utp-C module with the nascent 35S pre-rRNA requires prior loading of Rrp5pCitation15,Citation22 and recruitment of Pwp2/Utp-B is essential for the integration of U3 snoRNPCitation19 as well as Bms1p and Utp20p.Citation22 Bms1p and Utp20p loading is next required for the recruitment of the Imp3p-Imp4p-Mpp10p module and a subset of proteins to the nascent pre-ribosomes.Citation22
Interestingly, it has recently been proposed that proper assembly of SSU processome is monitored by a nucleolar surveillance mechanism involving TRAMP5 and exosome complexes to eliminate the RNA component of aberrant preribosomes.Citation17
It is predicted that the SSU processome interacts with other proteins in order for the cleavages of the pre-rRNA to occur.Citation14 To identify novel components in early step of ribosome biogenesis that might have been overlooked in genetic and mass-spectrometry analysis, we characterized YGR251W, a gene coding a protein localized in the nucleolus.Citation23 Using a microarray monitoring abundance of noncoding RNAs, Peng et al. revealed that expression of YGR251W is required for normal accumulation of mature 18S rRNA.Citation24 Here we present evidence that the yeast open reading frame YGR251W, we renamed Nop19, encodes for a ribosome biogenesis factor essential for cell viability. We demonstrate its association with 35S pre-rRNA and U3 snoRNA. Furthermore, we show that genetic depletion of Nop19p leads to accumulation of the 35S pre-rRNA as well as the aberrant 23S, and subsequent decrease in the levels of 20S pre-rRNA and mature 18S rRNA. These data indicate that Nop19p is a component of 90S preribosomal particle required for proper cleavages at sites A0, A1 and A2 in the pre-rRNA. Searches for tight protein-protein interactions with Nop19p within the 90S preribosome identified the DEAH-box RNA helicase Dhr2p and the U3 processome component Utp25p, both required for A0, A1 and A2 cleavages. Gradient analysis suggested that Nop19p is essential for the incorporation of Utp25p in preribosomes and the release of Dhr2p from preribosomes.
Results
Nop19p is a nucleolar protein associated with preribosomes.
A large number of ribosomal assembly factors have been identified by combining genetic studies and pre-ribosomal analysis using mass-spectrometry. To unveil protein that might have been overlooked in previous study, we focus our analysis on nucleolar proteins of unknown function. Previous global analyses of yeast proteins localization have shown that the protein Nop19p, encoded by the open reading frame YGR251W, localizes to the nucleolus. In order to confirm the subcellular localization of Nop19pCitation23, a construct expressing a C-terminal YFP fusion was integrated at the endogenous locus using a one step PCR strategy. Clones were verified by PCR and transformed with plasmid pUN100-mCherry-NOP1Citation25 expressing a mCherry-fused version of Nop1p (mCherry-Nop1p), a core component of box C/D snoRNPs involved in early, nucleolar stages of the pre-rRNA maturation pathway, to visualize the nucleolus. The strain was grown exponentially and living cells were analyzed by fluorescence microscopy (). The YFP signal observed in these conditions is highly enriched within a crescent-shaped region also containing mCherry-Nop1p, consistent with a nucleolar localization. Nop19p-YFP is also present at low levels in a region immediately adjacent to the nucleolus corresponding to the nucleoplasm. We concluded that Nop19p accumulates predominantly in yeast cell nucleoli suggesting that Nop19p might be a component of preribosomes. To get insights into the nature of macromolecular complexes containing Nop19p, we analyzed the sedimentation profile of Nop19p with that of (pre)ribosomal particles on density gradients. We used a strain expressing a C-terminal fusion between NOP19 and the TAP tag from the genomic locus (Open Biosystems). Expression of this fusion protein remained under the control of the endogenous promoter. Growth of the tagged strain was indistinguishable from wild-type strain (data not shown), indicating that the fusion protein is functional. Sedimentation profile of Nop19-TAP was tested by fractionation of a cell lysate on a 4.5%–45% sucrose gradient (). Western blot analysis of gradient fractions showed that Nop19p concentrates in two broad peaks. The dense fraction containing Nop19p-TAP (fractions 11–13) likely corresponds to the SSU processome/90S pre-ribosome whereas the light sedimenting fractions (fractions 1–3) probably represent the soluble protein plus UTP subcomplex(es). Signal was also observed in fractions 7–10 suggesting that few amounts of Nop19p are also present in pre-40S and/or pre-60S particles. Thus, a large part of Nop19p-TAP is engaged within large complexes, the density of which is consistent with that of various preribosomal particles. We next carried out immunoprecipitation experiments to assess whether Nop19p interacts physically with preribosomal particles. The NOP19::TAP strain was grown in the presence of glucose, and Nop19p-TAP was precipitated in non-denaturing conditions from a total cellular extract by the use of IgG-conjugated Sepharose. Following immunoprecipitations, bound RNAs were analyzed by northern hybridization () and compared with RNAs recovered in parallel from the non-tagged control strain (no TAG lanes). We found that the RNA component of 90S preribosomal particles, the 35S pre-rRNA is efficiently precipitated (∼20%) as well as the 23S pre-rRNA (∼15%), compared to the isogenic non tagged strain ( and lanes 1–4). The tight association of Nop19p with 23S pre-rRNA might suggests that A0, A1 and A2 cleavages are required for the release of the protein from pre-ribosomal particles. Late 20S and 27S pre-rRNAs were also co-precipitated with Nop19p-TAP clearly above background levels but appeared weaker than for 35S or 23S pre-rRNAs (respectively ∼4% and ∼2%), suggesting Nop19p is quickly released from these particles after A0, A1 and A2 cleavages. Our results show that, Nop19p is tightly associated with early pre-rRNA, component of the 90S preribosomal particles, a conclusion further supported by the presence, above the background, in the Nop19p-TAP immunoprecipitate, of various C/D and H/ACA snoRNAs ( and lanes 5–8).
Nop19p is required for cleavages of the pre-rRNA at sites A0, A1 and A2.
The YGR251W/NOP19 ORF was initially reported to be essential for cell viability.Citation26 In order to investigate the function of Nop19p in ribosome biogenesis, we constructed a yeast strain that conditionally expresses the Nop19p protein fused to the 3HA tag at the N-terminus (3HA-Nop19p protein), allowing its easy detection. The NOP19 open reading frame was tagged by the HA-encoding sequence and placed under the control of the regulated GAL1-10 promoter by homologous recombination,Citation27 creating strain GAL1::3HA::NOP19. This strain was propagated in a medium containing galactose as carbon source and was then shifted to a glucose-containing medium to allow depletion of Nop19p.
On galactose containing medium, the growth rate of GAL1::3HA::NOP19 strain and otherwise isogenic wild type were almost identical. Two hours after transfer to the non-permissive glucose medium, the growth rate of GAL1::3HA::NOP19 strain was already substantially reduced compared to wild-type, with a doubling time of ∼5 h. Growth essentially ceased by 25 h after transfer ().
Aliquots of GAL1::3HA::NOP19 cells grown in galactose-containing medium or grown for 1, 3, 6, 12, 24 and 48 hours in glucose-containing medium were harvested. From these aliquots, total proteins and RNAs were extracted and the kinetics of Nop19p depletion was assessed by western-blotting () analyses. In these conditions, the abundance of 3HA-Nop19p was strongly reduced after transfer to glucose medium and became undetectable after 1 h.
As a first step in assessing whether Nop19p is involved in the production of ribosomal subunits, we compared the ribosome profiles in the presence and absence of Nop19p on sucrose gradient (). GAL1::3HA::NOP19 and otherwise isogenic wild type cells were grown for 6 h in glucose-containing medium and sedimentation profiles were compared by 4.5% to 45% sucrose gradient analysis. Depletion of 3HA-Nop19p resulted in a clear reduction of the sedimentation peak corresponding to the free 40S subunits (fractions 7 and 8), a correlated strong increase of the 60S subunit sedimentation peak (fraction 10) and a concomitant polysome decrease (fractions 13–18). This phenotype clearly suggests a putative role of Nop19p in the production of the small ribosomal subunit.
To investigate the origin of this phenotype, we compared pre-rRNA processing in WT and Nop19p-depleted cells by northern-blot analysis (). Strains depleted of Nop19p showed defects in the pathway of 40S subunit synthesis. As early as 1 h after the nutritional shift, levels of the 35S pre-rRNA increased, while the 27SA2 and 20S pre-rRNAs were reduced ( and lanes 7–12). This suggests that early cleavages of 35S pre-rRNA at sites A0, A1 and A2, necessary for the production of 20S pre-rRNA (the immediate precursor to 18S rRNA) are somewhat impaired (for a cartoon of the pre-ribosomal RNA processing pathway, see ). This conclusion was supported by a strong accumulation of the aberrant 23S RNA, which is produced by direct cleavage of the 35S pre-rRNA at site A3 in the absence of cleavage at sites A0, A1 and A2. At later time points, the mature 18S rRNA was depleted in the strains lacking Nop19p, whereas levels of the 25S and 5.8S rRNAs were stable. According to phosphorimager quantifications, the level of 18S rRNA, relative to that of 25S rRNA, is diminished by 90% after 24 hours of growth in glucose-containing medium. The levels of the 27SB and 7S pre-rRNA were also lower in Nop19p-depleted cells after 12 h of depletion, but this may largely reflect reduced synthesis as a consequence of growth inhibition. The ratio between the long and short forms of 5.8S rRNA was unaltered, indicating that the alternative pre-rRNA processing pathways that generate these rRNAs both remain active. Together these observations indicate that depletion of Nop19p leads to the inhibition of cleavage at sites A0, A1 and A2.
The effects of reduced 3HA-Nop19p level on pre-rRNA processing were also assessed by pulse-chase labeling, performed 3 h after transfer of the GAL1::3HA::NOP19 strain to glucose medium. Consistent with the northern data, processing of the 35S pre-rRNA was delayed, whereas synthesis of 27SA and 20S pre-rRNA was greatly reduced in the strains depleted of Nop19p ( and lanes 8–14) compared to the wild type ( and lanes 1–7). Maturation of the 27SB pre-rRNA appeared to be slowed, but accumulation of mature 25S rRNA was not clearly reduced. Similar result was previously observed during Has1p depletion.Citation28 In contrast, 18S rRNA synthesis was strongly inhibited, most probably as a direct result of reduced 20S pre-rRNA production detected by this approach. These results confirmed that cleavages at A0, A1 and A2 are strongly impaired in cells lacking Nop19p.
Nop19p is not required for proper assembly of U3 processome.
As previously shown, Nop19p is a novel component of the 90S preribosome and is required for early cleavages of the 35S pre-rRNA. In order to get insight into the function of Nop19p in the hierarchical assembly of the 90S preribosome, we first determined whether Nop19p is required for proper accumulation of components of this particle. For this purpose, we individually expressed TAP-tagged versions of UTP proteins Utp17p (UTP-A), Pwp2p (UTP-B), Utp22p (UTP-C) as well as Rrp5p in the GAL1::3HA::NOP19 strain. Depletion of 3HA-Nop19p clearly did not result in reduced levels of any of these factors (data not shown), suggesting Nop19p is not required for their accumulation.
We next determined whether Nop19p is required for the incorporation of the UTP-A, UTP-B, UTP-C modules as well as Rrp5p within the SSU processome. To study the importance of Nop19p in the assembly of the 90S preribosome, we compared the sedimentation profiles of components of the UTP-A, UTP-B or UTP-C subcomplexes in WT and Nop19p-depleted cells (). For this experiment, we choose to deplete GAL1::3HA::NOP19 strains for 3 h. At this time point indeed, 3HA-Nop19p levels are already undetectable but ribosome biogenesis is only slightly affected, arguing against indirect effect. The A254 absorbance profiles and notably the large versus small ribosomal subunit ratio presented in the parts in confirmed that depletion of Nop19p induced a berely detectable defect in the production of the mature 40S ribosomal subunit. The Utp17p-TAP, Pwp2p-TAP, Utp22p-TAP or Rrp5-TAP proteins were still detected in fractions of the gradient containing high-molecular-weight complexes and none of them was mainly detected in the top fractions containing small particles and free proteins. We concluded that the preribosomal particles lacking Nop19p contain the tested components of the different UTP modules and that Nop19p is therefore probably not required for their incorporation.
We next studied whether Utp17p (UTP-A complex), Pwp2p (UTP-B complex), or Rrp5p are required for Nop19p recruitment to the nascent preribosomal particles. We replaced the endogenous promoters of the UTP17, PWP2 or RRP5 genes with the conditional GAL1 promoter in the previously described NOP19::TAP strain. These strains were shifted from a galactose- to a glucose-containing medium and grown to deplete Utp17p, Pwp2p or Rrp5p. Depletion of these factors did not result in a reduction of 3HA-Nop19p (data not shown), suggesting they are not required for Nop19p-TAP accumulation.
Extracts prepared from the depleted cells were analyzed by sedimentation on sucrose gradients. The absorbance profiles observed in Utp17p-, Pwp2p- and Rrp5p-depleted cells () are consistent with those reported previously by others in reference Citation15, Citation19 and Citation29 and very similar to those observed in the absence of Nop19p. In absence of Utp17p, Pwp2p and Rrp5p, Nop19p was still detected in the fractions of the gradient containing preribosomal particles. However, the sedimentation profiles of Nop19-TAP in Utp17p-, Pwp2p- and Rrp5p-depleted cells appeared significantly different from that observed with WT cells. The protein is indeed more concentrated in fractions 11 and 12 in WT cells whereas it appeared shifted to fractions 8 to 10 in depleted cells and appeared less concentrated in the top fractions containing free proteins and other low-molecular-weight complexes. These data indicate that the incorporation of Nop19p into preribosomes is not prevented in the absence of UTP-A or UTP-B components or Rrp5p. The observed shift in sedimentation may reflect that Nop19p remains associated with stalled preribosomes containing the aberrant 23S pre-rRNA. Therefore, Nop19p is essential for rRNA processing, but has no detectable function in hierarchical assembly of the 90S preribosome.
Dhr2p and Utp25p are in tight association with Nop19p.
In order to get insights into the function of this novel factor, we next attempted to identify “preferential” Nop19p-associated factors. The aim was to identify tight interactions between Nop19p and 90S preribosomal proteins using stringent conditions. Hence, Nop19p fused to the TAP tag (Nop19p-TAP) was purified over an IgG-sepharose column in native conditions. Nop19p-TAP-containing complexes bound to the column were then submitted to a “disruption treatment” consisting in extensive washes with buffers containing 200 mM, 1 M or 2 M salt (KCl) concentration (see Materials and Methods). The protein A-IgG interaction is resistant to 1 M and 2 M salt treatment,Citation30 but most of the proteins component of the 90S preribosome are expected to be released following these stringent washes. Following TEV elution, a second calmodulin affinity column was employed as described in reference Citation31. The polypeptides copurified with Nop19p following this modified TAP tag protocol were resolved by SDS-PAGE and stained with coomassie blue (Fig. S1). Coomassie-stained bands were excised and polypeptides contained in these bands were subjected to in-gel trypsin digestion. The resulting peptides were analyzed by on-line capillary liquid chromatography/nanospray ion trap tandem mass spectrometry, allowing identification of the proteins from which they were derived.
Among the identified factors, we noticed the presence of two 90S components, Dhr2p and Utp25p. Dhr2p is a DEAH-box RNA helicase essential for cell viability. Dhr2p is required for A0, A1 and A2 cleavages and so on for 18S rRNA production.Citation32 Dhr2p presents a strong conservation in its core domain with Dhr1p, one other DEAH-box RNA helicase also required for early 35S pre-rRNA cleavages. Moreover, Dhr2p has been shown to be associated in vivo with U3 processome components including U3 snoRNA, Mpp10p,Citation33 Utp22pCitation34 and Nop9p.Citation35 Utp25p is an essential factor, also required for A0, A1 and A2 cleavages (reviewed in ref. Citation36 and Citation37 and Fig. S2). As Dhr2p, Utp25p coimmunoprecipitated SSU processome factors such as U3 snoRNA, Mpp10p, Utp3p/Sas10p, Utp8p, Utp18p and Utp21p.Citation34,Citation36,Citation37 All together, these results suggested that Nop19p is a component of the SSU processome interacting “preferentially” with Dhr2p and Utp25p.
In order to independently verify the interactions seen using the modified TAP protocol, we individually expressed in the NOP19::TAP strain, HA-tagged versions of Dhr2p, Dhr1p and Utp25p as well as various UTP proteins including Utp17p (UTP-A), Pwp2p (UTP-B), Utp22p (UTP-C) and Rrp5p. Strains were exponentially grown in YPD medium at 30°C and cell pellets were broken in liquid nitrogen. Nop19p-TAPcontaining complexes bound to the IgG sepharose column were extensively washed with buffers containing 0.2 M, 1 M or 2 M KCl. Protein complexes were next eluted using a denaturing buffer. The released peptides were analyzed by western-blot using anti-HA antibodies (). As expected, all tested factors were found associated with Nop19p-TAP at low salt concentration (0.2 M). In accordance with our previous observations, Dhr2p-HA and Utp25p-HA interactions with Nop19p-TAP were resistant to high salt concentrations (1 M and 2 M) (). In contrast, Dhr1p-HA is totally released from the column after 1 M KCl washes. These results confirm that Nop19p stably interacts with Dhr2p-HA and Utp25p-HA when compared with other 90S preribosomal proteins. Moreover, little amounts of Utp17p-HA (UTP-A) and Pwp2p-HA (UTP-B) remain associated with Nop19p after high salt disruption, suggesting stable association of these two factors with Nop19p-TAP, althought to a lesser extend as compared with Dhr2p and Utp25p. UTP-C factor (Utp22p-HA), as well as Rrp5p-HA are still associated to Nop19p-TAP at 1 M salt concentration but are released from the column during 2 M KCl washes. All together, these results suggest that Nop19p is specifically and stably associated to Dhr2p and Utp25p. Moreover, stable association between Nop19p and UTP-A and UTP-B components were identified. Nop19p also appeared associated with UTP-C subcomplex and Rrp5p, to a lower extent however.
We next determined whether Nop19p is required for the incorporation of Dhr2p and Utp25p within the SSU processome. For this purpose, we compared the sedimentation profiles of Dhr2p-TAP and Utp25p-TAP in WT and Nop19p-depleted cells (). Western blot analysis of gradient fractions showed that Dhr2p concentrates in two broad peaks in WT cells. The lower peak of Dhr2p-TAP (fractions 11–14) may correspond to the SSU processome/90S preribosome whereas the slower sedimenting fractions (fractions 1–3) probably represents the free protein plus small protein complex(es). Signal is also observed in fractions 8–10 suggesting few amounts of Dhr2p are present in pre-40S and/or pre-60S particles. In Nop19p-depleted cells, Dhr2p was still present in heavy fractions, suggesting Nop19p is not required for its incorporation into the preribosomes. Interestingly, Dhr2p was no more detected in top fractions while it concomitantly accumulated in fractions 7–9. These results probably reflect that in absence of Nop19p, Dhr2p is trapped within stalled preribosomes containing the aberrant 23S pre-rRNA avoiding its recycling.
In accordance with observations by other,Citation37 Utp25p is present in high molecular mass fractions probably corresponding to SSU processome/90S and pre-40S preribosomes (). These results were also assessed by co-immunoprecipitation experiments which confirmed that Utp25p is associated with both the 35S and 20S pre-rRNAs (Fig. S2) data not shown. In absence of Nop19p, very weak signal was observed in high molecular complexes whereas signal strongly increased in top fractions. This data suggest that the incorporation of Utp25p into preribosomes is affected in absence of Nop19p. In these conditions, Utp25p accumulates in the pool of free proteins and/or small protein complexes.
Discussion
The assembly and maturation of preribosomes mobilize numerous trans-acting factors and snoRNPs associating transiently with the preribosomal particles in the nucleolus, the nucleoplasm and the cytoplasm to promote the formation of translation-competent ribosomal subunits. Many components of early preribosomes have been identified. However, most of them are only partially characterized. The current challenges in the field are to exhaustively establish the list of these components and to understand their precise function in the assembly and maturation processes of preribosomal particles. We report here the characterization of the YGR251W gene we called NOP19, encoding a novel nucleolar protein required for early cleavages of the pre-rRNAs leading to the production of the 40S ribosomal subunit.
We have shown by coimmunoprecipitation experiments that Nop19p interacts physically with the RNA components of the 90S preribosomal particles including U3 snoRNA and to a lesser extent with pre-40S particles. Upon Nop19p depletion, we observed a rapid depletion of the mature 18S rRNA. This decrease is the consequence of a defect in A0, A1 and A2 cleavages as Nop19p depletion results in the accumulation of the 35S and the 23S pre-rRNA concomitantly with a loss of the 20S pre-rRNA, the direct precursor of the mature 18S rRNA. A striking feature of the coprecipitation data was the strong recovery of the aberrant 23S RNAs with Nop19p. This RNA is detectable in wild-type cells, but at very low levels, and is strongly enriched in the Nop19p-TAP immunoprecipitate. These data suggest that the association of Nop19p with the presumably defective preribosomes that contain the 23S, is more stable than the association with preribosomes that are maturing normally. We propose that A0, A1 and A2 cleavages are required for the release of Nop19p from pre-ribosomal particles.
Our results show that Nop19p is not required for the association of components of the tUTP/UTP-A (Utp17p), UTP-B (Pwp2p) and UTP-C (Utp22p) complexes as well as Rrp5p with the 90S preribosomes. These results indicate that these modules assemble independently from Nop19p. These data also suggest that Nop19p is likely not a component of the tUTP/UTP-A subcomplex, as the incorporation of which is a prerequisite for the recruitment of most subsequent modules. Moreover, tUTP/UTP-A components are believed to be required for the transcription of the pre-rRNA or stability of the nascent transcript.Citation16,Citation17 Pulse chase experiments clearly revealed robust ongoing rDNA transcription in Nop19p-depleted cells. All together, these results strongly suggest Nop19p is not a bona fide component of the tUTP/UTP-A subcomplex. Conversely, components of tUTP/UTP-A, UTP-B and UTP-C complexes as well as Rrp5p are not required for the association of Nop19p with the 90S preribosome. It therefore appeared unlikely that Nop19p is a component of UTP-C subcomplex as it is still incorporated in the particle in absence of Rrp5p. One possibility is that Nop19p could be a component of UTP-B subcomplex. This hypothesis is supported by our co-immunoprecipitation data. Pwp2-HA is indeed efficiently co-immunoprecipitated with Nop19-TAP, even after high salt concentration treatment. Alternatively, Nop19p may be recruited to the 90S preribosomes, independently of the assembly of UTP modules. Actually, less than 50% of the 90S preribosome proteins are assigned to UTP subcomplexes.Citation14
TAP experiments under stringent conditions identified stable interactions between Nop19p and both Dhr2p and Utp25p. These two factors are known to be part of the U3 processome.Citation14,Citation33,Citation36,Citation37 The fact that these interactions resist to 2 M salt washes after immunoprecipitation suggests that Nop19p interacts preferentially with Dhr2p and Utp25p.
Dhr2p is a DEAH-box RNA helicase required for A0, A1 and A2 cleavages and so on for 18S rRNA production.Citation32 It was often proposed that SSU processome RNA helicases such as Dhr2p probably interact transiently with preribosomes and thus are not always detectable in tandem affinity purified particles. This made up difficult to get insight into the precise function of this group of factors. However, Dhr2p has been shown to be associated in vivo with U3 processome components including U3 snoRNA, Mpp10p,Citation33 Utp22pCitation34 and Nop9p.Citation35 Here we have shown that Dhr2p is stably associated with Nop19p. Dhr2p is recruited on the 90S preribosomes and is probably released shortly after A0, A1 and A2 cleavages in WT cells. In absence of Nop19p, Dhr2p remains associated with some preribosomes, probably stalled pre-ribosomes containing the aberrant 23S pre-rRNA. This avoids its recycling since no more free Dhr2p can be detected in gradient analysis. One hypothesis is that Nop19p directly participates in the release of Dhr2p from preribosomes in WT cells. However, we cannot exclude that the absence of A0, A1 and A2 cleavages due to the absence of Nop19p may be the direct cause of Dhr2p retention on preribosomes.
Utp25p was shown to provide a connection between UTP-B component (Utp21p) and Mpp10p subcomplexes.Citation36 Our data identified strong interactions between Nop19p and both Utp25p and the UTP-B subcomplex (Pwp2p). All together these data suggest that Utp25p and Nop19p can act together to bridge UTP-B and Mpp10p subcomplexes. It was largely proposed that this type of inter-subunit interactions promotes spatial and temporal association of the various subcomplexes during ribosome biogenesis. Actually, such a role for Utp25p and Nop19p remains unclear as none of them are required for proper loading of UTP subcomplexes within the 90S preribosomes (reviewed in ref. Citation36 and and ). Alternatively, Utp25p and Nop19p may contribute to fine-tune positioning of UTP-B and Mpp10p subcomplexes one relative to the other. Gradient analysis revealed that Nop19p is required for proper association of Utp25p with preribosomes. It's still unclear whether prior association of both protein is required for their integration within the 90S preribosome or if Nop19p association to preribosomes is a prerequisite to Utp25p loading onto the particle.
Materials and Methods
Strains, media, plasmids and cloning.
Standard procedures were used for the propagation of yeast using YPD medium (1% yeast extract, 2% peptone and 2% glucose), YPG medium (1% yeast extract, 2% peptone and 2% galactose) or YNB medium (0.67% yeast nitrogen base, 0.5% (NH4)2SO4 and 2% glucose or galactose) supplemented with the required amino acids. Yeast strains used in this study were derivatives of S. cerevisiae S288C or BY4741, originating from the S288C background (MATa, his3Δ1 leu2Δ0 met15Δ0 ura3Δ0). NOP19::TAP, DHR2::TAP, DHR1::TAP, RRP5::TAP, UTP17::TAP, UTP22::TAP, PWP2::TAP, UTP25::TAP strains were purchased from Open Biosystems where insertion of the tandem affinity purification (TAP) cassette was selected using HIS3MX6 marker. The GAL1::3HA::NOP19 strain, expressing 3HA-Nop19p under the control of a GAL1 promoter was constructed as follows: a PCR cassette containing kanamycin resistance (kanR) selectable marker, GAL1 promoter and the 3HA-tag sequence was amplified by PCR from the plasmid pFA6a-KanMX6-PGAL13HACitation27 using primers 903 and 905 (). The PCR fragment was inserted by homologous recombination upstream of the chromosomal NOP19 open reading frame in BY4741 strain. Transformants were selected for resistance to kanamycin and screened by immunoblotting.
The DHR1::TAP, DHR2::TAP, UTP25::TAP, RRP5::TAP, UTP17::TAP, PWP2::TAP and UTP22::TAP strains expressing 3HA-Nop19p under the GAL1 promoter control were constructed as follows: a PCR cassette containing the kanR selectable marker, GAL1 promoter and the 3HA-tag sequence was amplified by PCR from plasmid pFA6a-KanMX6-PGAL1-3HACitation27 using primers 903 and 905 (). The PCR fragment was inserted by homologous recombination upstream of the chromosomal NOP19 open reading frame in DHR1::TAP, DHR2::TAP, UTP25::TAP, RRP5::TAP, UTP17::TAP, PWP2::TAP and UTP22::TAP strains respectively. The GAL1::3HA::RRP5 strain expressing Nop19p-TAP was constructed as follows: a PCR cassette containing the kanR selectable marker, GAL1 promoter and the 3HA-tag sequence was amplified by PCR from plasmid pFA6a-KanMX6-PGAL1-3HACitation27 using 912/913 primers (). The PCR fragment was inserted by homologous recombination upstream of the chromosomal RRP5 open reading frame in NOP19::TAP strain. Transformants were selected using G418 and screened by immunoblotting.
The Gal::UTP17 and Gal::PWP2 strains expressing Nop19p-TAP were constructed by one-step PCR using pFA6a-KanMX6-PGAL1 Citation27 amplified with OMG054/OMG055, OMG050/OMG051, respectively.Citation29 The PCR fragment was inserted by homologous recombination upstream of the chromosomal UTP17 or PWP2 open reading frame in NOP19::TAP strain. Transformants were selected for kanamycin resistance and histidine prototrophy and screened by immunoblotting.
The PWP2::3HA, UTP17::3HA, UTP22::3HA, DHR2::3HA, DHR1::3HA, RRP5::3HA, UTP25::3HA strains expressing Nop19p-TAP were constructed by a one-step PCR using pFA6a-3HA-KanMX6 Citation27 amplified by primer couples 914/915, 916/917, 918/919, 920/921, 922/923, 793/792, 924/926 respectively. The PCR fragments were inserted by homologous recombination downstream of the chromosomal PWP2, UTP17, UTP22, DHR2, DHR1, RRP5, UTP25 open reading frames respectively in NOP19::TAP strain. Transformants were selected for kanamycin resistance and histidine prototrophy and screened by immunoblotting.
The NOP19::YFP strain containing a plasmid pUN100 NOP1::mCherry was constructed as followed: a PCR cassette containing URA3 gene from K. lactis as selectable marker and YFP-tag sequence was amplified by PCR from the plasmid pFA6aYFP-URA using primers 700 and 701 (). The PCR fragment was inserted by homologous recombination downstream in NOP19::TAP strain using the Swap-tag method.Citation38 Transformants were selected for uracile prototrophy and screened by immunoblotting. In a second step, plasmid pUN100 NOP1::mCherryCitation39 was transformed in the resulting strain. For the complete yeast strains used in this study, see .
Fluorescence microscopy.
For fluorescence microscopy, cells were grown in YPD medium at 30°C to an OD600 of 0.6. Aliquots were collected, washed and resuspended in dextrose-containing YNB supplemented with the needed amino acids. After washing, the cells were mounted on a slide and observed in the fluorescence microscope IX-81 (Olympus) equipped with a polychrome V monochromator and a CoolSNAP HQ camera (Roper Industries). Digital pictures were processed using Photoshop software (CS3 version; Adobe).
Western analyses.
Proteins from total extracts, obtained from gradient fractions after TCA precipitation or from immunoprecipitated pellets were separated on 4–12% polyacrylamide/SDS gels (Invitrogen) and transferred to hybond-ECL membranes (GE Healthcare). Nop1p was detected as described in reference Citation40. TAP-tagged proteins were detected using rabbit PAP (Sigma) diluted 10,000 fold. HA-tagged proteins were detected using anti-HA peroxydase antibodies (Roche) diluted 1,000-fold.
Sucrose gradient sedimentation experiments.
Cells were grown exponentially and treated for 30 min with 50 µg/ml cycloheximide (Sigma) added directly to the culture medium. Cells were collected by centrifugation, frozen in liquid nitrogen and broken with in a mortar. Dry extracts were resuspended with approximately 1 volume of ice-cold A150 buffer (20 mM Tris-Cl [pH 8], 5 mM MgCl2, 0.2% Triton X100, 150 mM KCl) supplemented with 1 mM dithiothreitol, 1x Complete EDTA-free protease inhibitor cocktail (Roche), 0.5 mM PMSF and 50 µg/ml cycloheximide. Extracts were clarified by centrifugation at 13,000 rpm and 4°C for 10 min and quantified by measuring absorbance at 260 nm. About 10 A260 units were loaded on 4.5% to 45% (w/v) sucrose gradients in specicific buffer (50 mM Tris-Acetate [pH 7.5], 50 mM NH4Cl, 12 mM MgCl2). Gradients were centrifuged for 150 min at 39,000 rpm and 4°C in an Optima L-100XP Ultracentrifuge (Beckman-Coulter) using the SW41Ti rotor without brake. Following centrifugation, 17 fractions of approximately 600 µl (33 s) each were collected from the top of the gradients by use of a Foxy Jr. fraction collector (Teledyne ISCO). The absorbance at 254 nm was measured during collection with a UA-6 device (Teledyne ISCO).
RNA extractions and northern hybridizations.
RNA extractions and northern hybridizations were performed as previously described in reference Citation41. For high molecular weight RNA analysis, 2 µg of total of RNA were glyoxal denatured and resolved on a 1.2% agarose gel. Low molecular weight RNA products were resolved on 8% Polyacrylamide/8.3 M urea gels. Oligonucleotides used for northern hybridizations are listed in .
Immunoprecipitations.
Total cell extracts were prepared from strains expressing TAP-tagged proteins or untagged strains as control. Growing cells were frozen in liquid nitrogen and broken in a mortar. Immunoprecipitations and analysis of coprecipitated RNAs were performed as previously described in reference Citation42. Tot/IP ratios loaded were 1/10 for agarose and acrylamide gels.
Tandem affinity purifications.
Cell pellets (corresponding to about 5 × 1010 cells) frozen in liquid nitrogen were broken in a mortar and resuspended in A200 KCl buffer [20 mM Tris-Cl (pH 8.0), 5 mM MgAc, 200 mM KCl, 0.2% Triton X-100] supplemented with 1 mM dithiothreitol, 1x Complete EDTA-free protease inhibitor cocktail (Roche) and 0.5 U/µl RNasin (Promega).
For TAP purifications under native conditions. Cell extracts were clarified by centrifugation at 14,000 rpm (21,000x g) for 10 min at 4°C. Clarified extracts were incubated with 200 µl (bed volume) of IgG-Sepharose beads (GE Healthcare) for 4 h on a rocking table. Beads were split in 3 different columns and were extensively washed with A200, A1000 or A2000 buffers (A1000 corresponds to A200 buffer except 1 M KCl, A2000 corresponds to A200 buffer except 2 M KCl). Columns were next equilibrated with ice-cold TEV cleavage buffer (10 mM Tris-Cl [pH 8.0], 150 mM NaCl, 0.1% NP-40, 0.5 mM EDTA, 1 mM dithiothreitol). Beads were resuspended with 1 ml of TEV cleavage buffer and incubated for 2 h at 16°C on a rocking Table in the presence of 100 units of ActTEV protease (Invitrogen). Eluted samples (about 1 ml) were mixed with 3 ml of calmodulin binding buffer [10 mM Tris-Cl (pH 8.0), 150 mM NaCl, 1 mM MgAc, 1 mM imidazole, 2 mM CaCl2, 0.1% NP-40, 10 mM β-mercaptoethanol] and 3 µl of 1 M CaCl2 and incubated with 200 µl of calmodulin beads (Stratagene) at 4°C for 1 h on a rocking table. Beads were washed with 40 ml of calmodulin binding buffer and proteins were eluted by the addition of 6 × 200 µl of calmodulin elution buffer [10 mM Tris-Cl (pH 8.0), 150 mM NaCl, 1 mM MgAc, 1 mM imidazole, 2 mM EGTA, 0.1% NP-40, 10 mM β-mercaptoethanol]. Eluted proteins were precipitated with 20% trichloroacetic acid (TCA), rinsed with acetone and resuspended with 20 µl of 1x SDS gel-loading buffer [40 mM Tris-Cl (pH 6.8), 2% SDS, 10% glycerol, 25 mM dithiothreitol, 0.1% bromophenol blue]. Samples were loaded on 4–12% Bis-Tris gels. Gels were briefly (15 min) stained with silver staining solution (Fermentas), and pieces of gels containing the samples were excised. The proteins contained in these samples were analyzed by mass spectrometry as described in reference Citation43.
Pulse chase analysis.
Metabolic labeling of pre-rRNAs was performed as previously described in reference Citation44, with the following modifications. Strains were pre-grown in synthetic galactose medium lacking adenine and transferred to glucose medium lacking adenine for 3 h (GAL1::3HA::NOP19). Cultures at OD600 0.8 were labeled with [8-3H] adenine (TRK343, Amersham) for 2 min followed by a chase with excess cold adenine. 1 ml samples were collected 1, 2, 5, 10, 20 and 30 min following the addition of cold adenine and cell pellets were frozen in liquid nitrogen. RNAs were then extracted and precipitated with ethanol.
NanoLC-MS/MS and data analyses.
Gel bands were excised and proteins were subjected to in-gel reduction and alkylation followed by in-gel tryptic digestion using modified porcine trypsin (Promega, Lyon, France). Concentrated peptide extracts were dissolved in 15 µL of 0.05% TFA in 10% ACN and analyzed by online nanocapillary HPLC (Ultimate3000, Dionex) coupled to an LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). Peptides were separated on a 75 µm ID × 15 cm C18 column (packed in-house with Reprosil C18-AQ Pur 3 µm resin, Dr. Maisch; Proxeon Biosystems, Odense, Denmark) after loading onto a 300 µm ID × 5 mm PepMap C18 precolumn (Dionex). Peptides were eluted using a 5 to 50% linear gradient of solvent B in 55 min (solvent A was 0.2% formic acid in 5% ACN and solvent B was 0.2% formic acid in 80% ACN). Data were acquired in data-dependent scan using a 60 s dynamic exclusion window. MS scans over the 300–2,000 m/z range were followed by MS/MS scans of the five most intense ions. The Mascot Daemon software (version 2.2.0.3, Matrix Science) was used to perform database searches in batch mode with all the raw files acquired against Saccharomyces cerevisiae Swiss-Prot TrEMBL database (UniProt 15.5). Mascot results were parsed with the in-house developed software Mascot File Parsing and Quantification (MFPaQ) version 4.0.0,Citation45,Citation46 and an identified protein was considered a hit if it was identified with at least two peptides with a score greater than the significance threshold score for a probability p < 0.05 or at least one pep-tide with a score greater than the significance threshold score for p < 0.001, as determined by the Mascot Search program.
Specific protein partners of Nop19p (YGR251w) were identified by a differential semi-quantitative analysis with the control (Saccharomyces cerevisiae BY4741) identified proteins using the MS/MS spectral counting method adapted from Liu et al.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Figures and Tables
Figure 1 Scheme of the pre-rRNA processing pathway in S. cerevisiae. (A) Initial 35S pre-rRNA precursor with detailed cleavages sites. (B) Endonucleolytic and exonucleolytic cleavages leading to the production of mature 18S, 5.8S and 25S (reviewed in ref. Citation22).
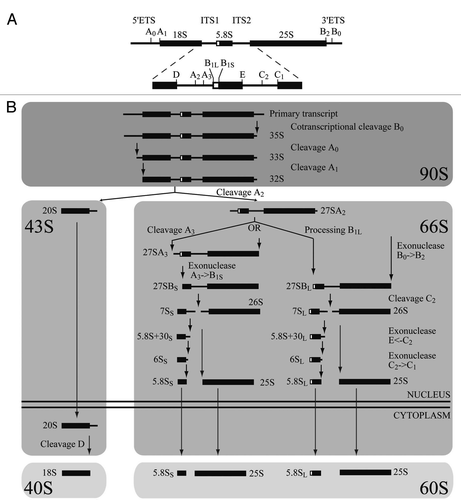
Figure 2 Nop19p is a nucleolar protein associated with preribosomes. (A) Subcellular localization of Nop19p. Yeast strain expressing Nop19p-YFP and mCherry-Nop1p were grown exponentially and cell samples were used for fluorescence microscopy analysis. (B) Sedimentation profile of Nop19p on a sucrose gradient. A total extract prepared from NOP19::TAP cells growing exponentially was sedimented through a sucrose gradient and 17 fractions were collected. The corresponding A254 profile is displayed with the characteristic annotated peaks. Each fraction was TCA precipitated and Nop19-TAP was detected by western blotting using PAP antibodies. (C) Nop19p co-immunoprecipitates with 35S and 23S pre-rRNAs. Northern-blot analysis of (pre-)rRNAs coprecipitated with TAP-tagged version of Nop19p (lanes 3–4 and 7–8) or from control experiments using extracts of cells lacking a tagged protein (lanes 1, 2, 5 and 6). Immunoprecipitation was performed on cell extracts using IgG-Sepharose. RNAs were extracted from the pellet after precipitation (lanes IP) or from total cell extract (lanes Tot) corresponding to 10% of the input used for the immunoprecipitation reactions. Following separation, RNAs were transferred to a nylon membrane and hybridized with anti-sense oligonucleotides corresponding to various (pre-) rRNAS and snoRNAs.
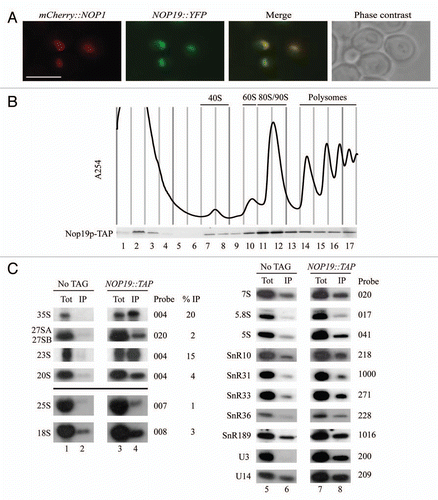
Figure 3 Nop19p depletion affects 40S ribosomal subunit accumulation in yeast cells. (A) Growth rate of wild-type and Gal::3HA::NOP19 strains following a transfer from permissive galactose medium to glucose medium for the times indicated. Cells were maintained in exponential growth throughout the time course by dilution into pre-warmed medium. (B) Western-blot analysis of 3HA-Nop19p depletion. Total proteins were extracted at the times indicated and analyzed by western blot. Accumulation of 3HA-Nop19p and Nop1p was respectively detected using anti-HA and Nop1p-specific antibodies. (C) Ribosome profiles in Nop19p-depleted cells. Gal::3HA::NOP19 and WT BY4741 strains were grown up to 0.6 (OD600) on galactose medium and shifted to glucose for 6 h. Total cell extracts were prepared and centrifuges through 4.5% to 45% sucrose gradients and 17 fractions were collected. The A254 absorbance profile is presented.
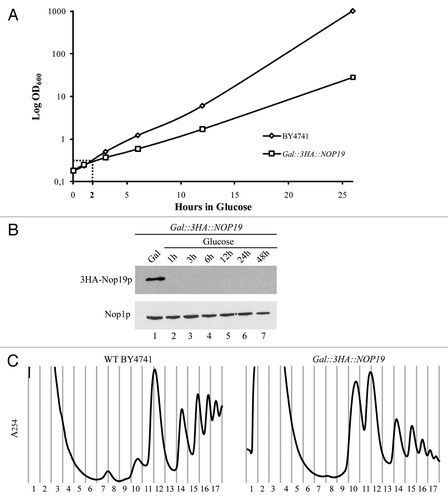
Figure 4 Nop19p depletion leads to a defect in A0, A1 and A2 cleavages. (A) WT BY4741 and Gal::3HA::NOP19 strains were shifted from a galactose to a glucose medium. Samples were collected before and at different times after the nutritional shift. Total RNAs were extracted from these cell samples, and the accumulation of the different pre-rRNAs, rRNAs and sn(o)RNAs was analyzed by northern-blot. (B) Pulse-chase labeling of RNAs. WT BY4741 and Gal::3HA::NOP19 cells were grown in galactose containing medium and were next shifted in glucose for 3 h. Cells were then pulse labeled with [8-3H] adenine for 2 min. Samples were collected 0, 1, 2, 5, 10, 20 and 30 min after addition of an excess of cold adenine. Total RNAs were extracted from these samples, separated by gel electrophoresis and transferred to a nylon membrane.
![Figure 4 Nop19p depletion leads to a defect in A0, A1 and A2 cleavages. (A) WT BY4741 and Gal::3HA::NOP19 strains were shifted from a galactose to a glucose medium. Samples were collected before and at different times after the nutritional shift. Total RNAs were extracted from these cell samples, and the accumulation of the different pre-rRNAs, rRNAs and sn(o)RNAs was analyzed by northern-blot. (B) Pulse-chase labeling of RNAs. WT BY4741 and Gal::3HA::NOP19 cells were grown in galactose containing medium and were next shifted in glucose for 3 h. Cells were then pulse labeled with [8-3H] adenine for 2 min. Samples were collected 0, 1, 2, 5, 10, 20 and 30 min after addition of an excess of cold adenine. Total RNAs were extracted from these samples, separated by gel electrophoresis and transferred to a nylon membrane.](/cms/asset/5893409e-992e-4afb-b649-a304fef36515/krnb_a_10917699_f0004.gif)
Figure 5 Nop19p and U3 processome assembly. (A) Depletion of Nop19p in yeast does not affect the incorporation of components of the UTP-A, UTP-B or UTP-C modules within preribosomes. Strains expressing TAP-tagged versions of Utp17p, Pwp2p, Utp22p or Rrp5p that were otherwise WT or expressing 3HA-Nop19p under the control of the GAL1 promoter were transferred from galactose- to glucose-based medium and grown for 3 h. Total extracts prepared from these cells were sedimented through sucrose gradients. Western blot experiments were carried out using rabbit PAP to detect TAP-tagged proteins in the different fractions. (B) Sedimentation profile of Nop19-TAP in WT cells (upper left part) or in cells lacking Utp17p (upper right part), Pwp2p (lower left part) or Rrp5p (lower right part). The GAL1::UTP17, GAL1::PWP2 and GAL1::RRP5 strains expressing Nop19-TAP were shifted from galactose- to glucose-containing medium and grown for 14 h, to deplete the corresponding proteins. As a control, the Nop19::TAP strain grown in the presence of glucose was used. Total extracts prepared from these cell samples were sedimented through sucrose gradients. The proteins contained in each fraction were analyzed by western blotting using PAP antibodies to detect Nop19-TAP.
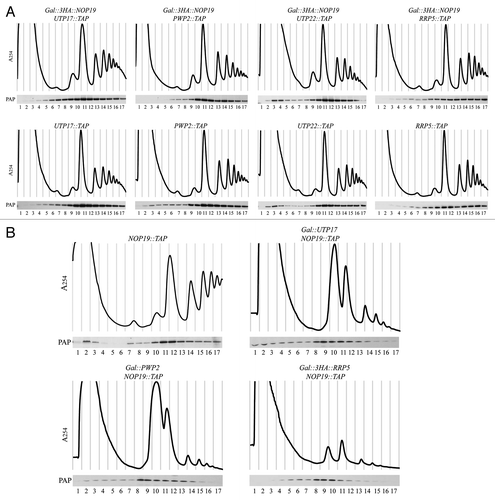
Figure 6 TAP purification of Nop19p-TAP under stringent conditions. (A) Nop19p-TAP was affinity purified under native conditions (see Materials and Methods). Nop19p-TAP-containing complexes bound to IgG were extensively washed with buffers containing 200 mM, 1 M or 2 M salt (KCl) concentration before Tev elution. Final purified samples were separated by SDS-polyacrylamide gel electrophoresis and observed after silver staining coloration. Samples were next subjected to mass spectrometry analysis. (B) Proteins specifically associated with Nop19p. Interactions identified by mass spectrometry analysis were individually verified. 3HA-tag versions of proteins of interest (Dhr2p, Utp25p, Dhr1p, Utp17p, Pwp2p, Utp22p, Rrp5p) were expressed in a NOP19::TAP background strain. Total extracts were immobilized on IgG and submitted to extensive washes with buffers containing 200 mM, 1 M or 2 M salt (KCl) before elution. Eluted proteins were analyzed by western blotting using PAP and HA antibodies.
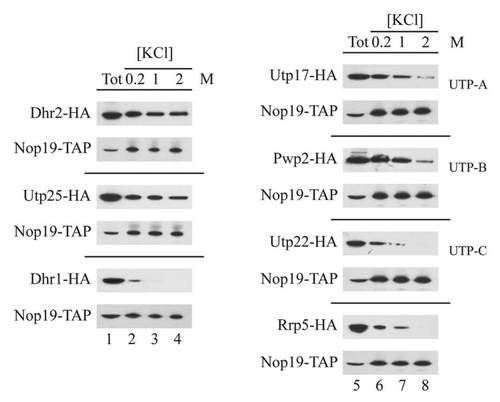
Figure 7 Depletion of Nop19p affects the sedimentation profile of Dhr2p and Utp25p. Strains expressing TAP-tagged versions of Dhr2p (A) and Utp25p (B) that were otherwise WT or expressing 3HA-Nop19p under the control of the GAL1 promoter were transferred from galactose- to glucose-based medium and grown for 3 h. Total extracts prepared from these cells were sedimented through sucrose gradients. Western blot experiments were carried out using rabbit PAP to detect TAP-tagged proteins in the different fractions.
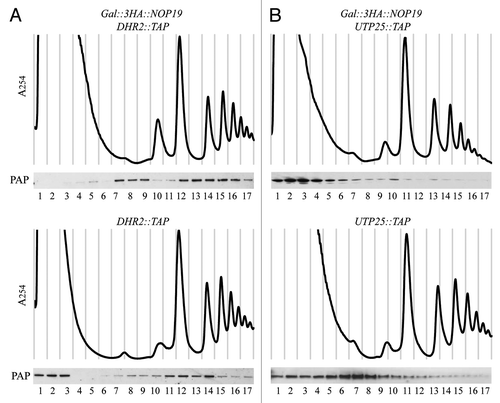
Table 1 List of oligonucleotides used in this study
Table 2 List of yeast strains used in this study
Additional material
Download Zip (7.9 MB)Acknowledgements
We are very grateful to Anthony Henras for critical reading of the manuscript. We acknowledge members of the GADAL lab for help, advice and discussion. We are also grateful to Jorge Pérez-Fernandez for helpful discussions and protocols concerning gradient experiments. We thank Laura Castillo and Justine Constans for their involvement in this project and David Villa for help with figures. This work also benefited of the assistance of the electron microscopy facility of the Institut Fédératif de Recherche 109 and of the imaging platform of Toulouse TRI. This work was supported by an Action Thématique et Incitative sur Programme (ATIP) Jeunes Chercheurs grant from Centre National de la Recherche Scientifique, by Agence Nationale de la Recherche (Nucleopol, ODynRib and Ribeuc programme), and Jeune équipe from Fondation pour la recherche médicale (FRM). E.C. is supported by a Ph.D. fellowship from the Ministère de l'Education Nationale, de l'Enseignement Supérieur et de la Recherche.
References
- Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Ann Rev Gen 1999; 33:261 - 311
- Lafontaine DL, Tollervey D. The function and synthesis of ribosomes. Nat Rev Mol Cell Biol 2001; 2:514 - 520
- de la Cruz J, Kressler D, Linder P. Olson MOJ. Ribosomal subunit assembly. The Nucleolus 2003; KluwerAcademic/Plenum Publishers 262 - 290
- Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene 2003; 313:17 - 42
- Dez C, Tollervey D. Ribosome synthesis meets the cell cycle. Curr Opin Microbiol 2004; 7:631 - 637
- Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, et al. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci 2008; 65:2334 - 2359
- Zhang J, Harnpicharnchai P, Jakovljevic J, Tang L, Guo Y, Oeffinger M, et al. Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev 2007; 21:2580 - 2592
- Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J 2001; 20:3617 - 3622
- Bachellerie JP, Cavaille J, Huttenhofer A. The expanding snoRNA world. Biochimie 2002; 84:775 - 790
- Terns MP, Terns RM. Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin. Gene Expr 2002; 10:17 - 39
- Dragon F, Gallagher JE, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 2002; 417:967 - 970
- Bernstein KA, Gallagher JE, Mitchell BM, Granneman S, Baserga SJ. The small-subunit processome is a ribosome assembly intermediate. Eukaryot Cell 2004; 3:1619 - 1626
- Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, et al. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol Cell 2004; 16:943 - 954
- Phipps KR, Charette JM, Baserga SJ. The SSU Processome in Ribosome Biogenesis—Progress and Prospects. WIREs RNA 2011; 2:1 - 21
- Perez-Fernandez J, Roman A, De Las Rivas J, Bustelo XR, Dosil M. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol Cell Biol 2007; 27:5414 - 5429
- Gallagher JE, Dunbar DA, Granneman S, Mitchell BM, Osheim Y, Beyer AL, et al. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev 2004; 18:2506 - 2517
- Wery M, Ruidant S, Schillewaert S, Lepore N, Lafontaine DL. The nuclear poly(A) polymerase and Exosome cofactor Trf5 is recruited cotranscriptionally to nucleolar surveillance. RNA 2009; 15:406 - 419
- Segerstolpe A, Lundkvist P, Osheim YN, Beyer AL, Wieslander L. Mrd1p binds to pre-rRNA early during transcription independent of U3 snoRNA and is required for compaction of the pre-rRNA into small subunit processomes. Nucleic Acids Res 2008; 36:4364 - 4380
- Dosil M, Bustelo XR. Functional characterization of Pwp2, a WD family protein essential for the assembly of the 90 S pre-ribosomal particle. J Biol Chem 2004; 279:37385 - 37397
- Grandi P, Rybin V, Bassler J, Petfalski E, Strauss D, Marzioch M, et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol Cell 2002; 10:105 - 115
- Krogan NJ, Peng WT, Cagney G, Robinson MD, Haw R, Zhong G, et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol Cell 2004; 13:225 - 239
- Perez-Fernandez J, Martin-Marcos P, Dosil M. Elucidation of the assembly events required for the recruitment of Utp20, Imp4 and Bms1 onto nascent pre-ribosomes. Nucleic Acids Res 2011;
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, et al. Global analysis of protein localization in budding yeast. Nature 2003; 425:686 - 691
- Peng WT, Robinson MD, Mnaimneh S, Krogan NJ, Cagney G, Morris Q, et al. A panoramic view of yeast noncoding RNA processing. Cell 2003; 113:919 - 933
- Albert B, Leger-Silvestre I, Normand C, Ostermaier MK, Perez-Fernandez J, Panov KI, et al. RNA polymerase I-specific subunits promote polymerase clustering to enhance the rRNA gene transcription cycle. J Cell Biol 2011; 192:277 - 293
- Sartori G, Mazzotta G, Stocchetto S, Pavanello A, Carignani G. Inactivation of six genes from chromosomes VII and XIV of Saccharomyces cerevisiae and basic phenotypic analysis of the mutant strains. Yeast 2000; 16:255 - 265
- Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 1998; 14:953 - 961
- Emery B, de la Cruz J, Rocak S, Deloche O, Linder P. Has1p, a member of the DEAD-box family, is required for 40S ribosomal subunit biogenesis in Saccharomyces cerevisiae. Mol Microbiol 2004; 52:141 - 158
- Gerus M, Bonnart C, Caizergues-Ferrer M, Henry Y, Henras AK. Evolutionary Conserved Function of RRP36 in Early Cleavages of the Pre-ribosomal RNA and Production of the 40S Ribosomal Subunit. Mol Cell Biol 2009;
- Henras AK, Capeyrou R, Henry Y, Caizergues-Ferrer M. Cbf5p, the putative pseudouridine synthase of H/ACA-type snoRNPs, can form a complex with Gar1p and Nop10p in absence of Nhp2p and box H/ACA snoRNAs. RNA 2004; 10:1704 - 1712
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 1999; 17:1030 - 1032
- Colley A, Beggs JD, Tollervey D, Lafontaine DL. Dhr1p, a putative DEAH-box RNA helicase, is associated with the box C + D snoRNP U3. Mol Cell Biol 2000; 20:7238 - 7246
- Granneman S, Bernstein KA, Bleichert F, Baserga SJ. Comprehensive mutational analysis of yeast DEXD/H box RNA helicases required for small ribosomal subunit synthesis. Mol Cell Biol 2006; 26:1183 - 1194
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 2006; 440:637 - 643
- Thomson E, Rappsilber J, Tollervey D. Nop9 is an RNA binding protein present in pre-40S ribosomes and required for 18S rRNA synthesis in yeast. RNA 2007; 13:2165 - 2174
- Charette JM, Baserga SJ. The DEAD-box RNA helicase-like Utp25 is an SSU processome component. RNA 2010; 16:2156 - 2169
- Goldfeder MB, Oliveira CC. Utp25p, a nucleolar Saccharomyces cerevisiae protein, interacts with U3 snoRNP subunits and affects processing of the 35S pre-rRNA. FEBS J 2010; 277:2838 - 2852
- Sung MK, Ha CW, Huh WK. A vector system for efficient and economical switching of C-terminal epitope tags in Saccharomyces cerevisiae. Yeast 2008; 25:301 - 311
- Berger AB, Cabal GG, Fabre E, Duong T, Buc H, Nehrbass U, et al. High-resolution statistical mapping reveals gene territories in live yeast. Nat Methods 2008; 5:1031 - 1037
- Dez C, Henras A, Faucon B, Lafontaine D, Caizergues-Ferrer M, Henry Y. Stable expression in yeast of the mature form of human telomerase RNA depends on its association with the box H/ACA small nucleolar RNP proteins Cbf5p, Nhp2p and Nop10p. Nucleic Acids Res 2001; 29:598 - 603
- Beltrame M, Tollervey D. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J 1992; 11:1531 - 1542
- Dez C, Froment C, Noaillac-Depeyre J, Monsarrat B, Caizergues-Ferrer M, Henry Y. Npa1p, a component of very early pre-60S ribosomal particles, associates with a subset of small nucleolar RNPs required for peptidyl transferase center modification. Mol Cell Biol 2004; 24:6324 - 6337
- Lebaron S, Froment C, Fromont-Racine M, Rain JC, Monsarrat B, Caizergues-Ferrer M, et al. The splicing ATPase prp43p is a component of multiple preribosomal particles. Mol Cell Biol 2005; 25:9269 - 9282
- Tollervey D, Lehtonen H, Jansen R, Kern H, Hurt EC. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation and ribosome assembly. Cell 1993; 72:443 - 457
- Mouton-Barbosa E, Roux-Dalvai F, Bouyssie D, Berger F, Schmidt E, Righetti PG, et al. In-depth exploration of cerebrospinal fluid by combining peptide ligand library treatment and label-free protein quantification. Mol Cell Proteomics 9:1006 - 1021
- Bouyssie D, Gonzalez de Peredo A, Mouton E, Albigot R, Roussel L, Ortega N, et al. Mascot file parsing and quantification (MFPaQ), a new software to parse, validate and quantify proteomics data generated by ICAT and SILAC mass spectrometric analyses: application to the proteomics study of membrane proteins from primary human endothelial cells. Mol Cell Proteomics 2007; 6:1621 - 1637
- Liu H, Sadygov RG, Yates JR 3rd. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem 2004; 76:4193 - 4201