Abstract
RLIP76 (RalBP1) is a multidomain protein that is a downstream effector of the small GTPases RalA and RalB. As well as the Ral binding domain it contains a RhoGAP domain active against Cdc42 and Rac1. RLIP76 also binds to proteins involved in endocytosis and to R-Ras. We recently solved the structure of the Ral binding domain of RLIP76 and the structure of the complex that it forms with RalB. The structure shows that, unlike the other Ral effectors characterized so far, RLIP76 forms a coiled-coil that interacts with RalB. The RLIP76 Ral binding domain binds to both the switch regions of RalB, which are the parts of the G protein that chance conformation upon nucleotide exchange. Here, we review our structure and discuss how it sheds light on the other functions of RLIP76.
RLIP76 was discovered simultaneously by three different groups 15 years agoCitation1–Citation3 as a downstream effector for the Ral GTPases. It originally had three names: RLIP76, RalBP1 and RIP1, although the former two are now most commonly used. The discovery of RLIP76 attracted some interest because it was found that as well as containing a binding domain for RalA and RalB, it had a RhoGAP domain, suggesting that it provided a link between Ras and Rho family-controlled signaling pathways. GAP assays with the three best-characterized Rho proteins, RhoA, Cdc42 and Rac1, showed that RLIP76 has GAP activity for Rac1 and Cdc42 but not for RhoA, nor indeed had it any activity towards Ral, Ras or Rap1a.
Since its characterization as a Ral effector and RhoGAP, RLIP76 has been implicated in various interesting cellular events. Firstly, yeast two hybrid screens have shown that RLIP76 interacts with proteins that are involved in endocytosis. The N-terminal region of RLIP76 binds to the AP2 adaptor complex, which is involved in clathrin-mediated endocytosis from the plasma membrane,Citation4 while the C-terminal region of RLIP76 associates with the EH domain-containing proteins REPS1 and REPS2, which are phosphorylated in response to EGF stimulation and presumably involved in receptor-mediated endocytosis.Citation5,Citation6 More recently, an interaction was found between the Epsin N-terminal homology domain of Epsin and RLIP76,Citation7 although in this case it was suggested that this interactions leads to cell migration and invasion. RLIP76 is upregulated in bladderCitation8 and ovarianCitation9 cancers and intriguingly has been shown to act as a membrane transporter that pumps glutathione conjugates, including chemotherapeutics, out of cells.Citation10 Finally, RLIP76 has been shown to play a role in long-term depression in synapses, by binding to the post-synaptic scaffold protein PSD-95 upon RalA activation.Citation11
Despite its fascinating and important cellular roles, there was no structural information available at all for RLIP76 until this year when we solved the solution structure of the Ral binding domain (RBD) and the complex that this domain forms with RalB. The RBD itself forms a very simple structure, being a coiled-coil comprising two α-helices, each of about 20 amino acids. Although it is a coiled-coil, the isolated RBD does not show any propensity for dimerization in solution. Our experience with another coiled-coil system, the HR1b domain from PRK1 in complex with Rac1, showed that the effector interacted with the C-terminal polybasic region of the small GTPase.Citation12 We were therefore careful to ensure that the C-terminal regions and 11 residue N-terminal extension of RalB were unnecessary for a high affinity interaction with the RBD of RLIP76. The structure of the RLIP76-RalB complex shows that, as is often the case with small GTPases and their effectors, the two switch regions of the GTPase are involved in the binding surface (). The switch regions of the GTPase are those that change conformation dramatically when the nucleotide is exchanged and are therefore most sensitive to the activation state of the small GTPase (reviewed in ref. Citation13). In the case of RalB-RLIP76, both switch regions are involved in the interaction. In some GTPases, only switch 1, often known as the effector loop, is involved in the interaction with effectors, e.g., the canonical Rap1A-Raf1 complex.Citation14 Furthermore, the structure of the Ral effector Sec5 in complex with RalA shows that here also switch 1 is exclusively involved in the interaction.Citation15 It is interesting to note that when both switches interact with effectors, for example in the RLIP76-RalB complex and the Exo84-RalA complex, there tends to be a larger surface area buried in the interaction interface (usually just under 2,000 Å2) compared to those complexes where only switch 1 is involved (usually around 1,000 Å2). The different buried surface areas are not translated into different affinities, which are in the 10–300 nM range for most small GTPase effectors. The functional significance of using one or both switch regions for binding effectors is not yet clear but it is tempting to speculate that when effectors only contact switch 1, this would leave switch 2 available to mediate contacts with other proteins. For example, GAP proteins tend to interact strongly with switch 2, so some initial contacts could be formed by a GAP binding to switch 2, even though switch 1 is engaged by an effector. This could indicate that effector interactions that do not utilize switch 2 might activate pathways that require fast access by GAP proteins to downregulate the signal.
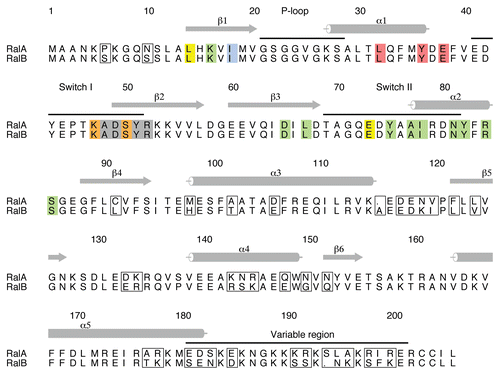
There are two Ral proteins in humans, RalA and RalB, which are 82% identical but which have been shown to have very different properties in tumor cells (reviewed in ref. Citation16): RalA is required for anchorage-independent proliferation, while RalB is necessary for suppression of apoptosis.Citation17 It is not clear which effectors are required for these dichotomous properties of the Ral proteins but it is striking that the sequences of RalA and RalB that interact with RLIP76, Sec5 and Exo84 are completely conserved () and concomitantly that Sec5 and RLIP76 bind to RalA and RalB with very similar affinities, at least in vitro.Citation15,Citation18,Citation19 This implies either that other effectors mediate the different roles of RalA and RalB or that the in vivo affinities do not reflect those that have been measured in vitro. The sequences of the Ral proteins diverge in the C-terminal half of the sequence, particularly in the last 20 residues (the ‘variable region’), indicating that regions outside the Sec5/Exo84/RLIP76-binding sites are responsible for the differential functions of RalA and RalB, either through differential localization, which would regulate effector availability or by mediating binding to as yet undefined effectors.
What are the effects of RLIP76 binding to Ral proteins? It has been shown that Ral binding has no effect on the ability of the RhoGAP domain to stimulate the GTPase activity of Cdc42 and Rac1 in vitro. There does however appear to be a stimulation of the RhoGAP activity upon binding to activated RalA in vivo,Citation20 perhaps suggesting that the consequence of the interaction for GAP activity is one of localization to the correct membrane. RLIP76 contains more than just a GAP domain juxtaposed to an RBD and there are other less well-characterized functions of RLIP76 that could be directly affected by Ral binding: RLIP76 has been shown to behave as a plasma membrane pump;Citation10 RalA, RLIP76 and POB1 regulate endocytosis of EGF and insulin receptors;Citation21 and RLIP76 binds Epsin to bring about cell migration and invasion.Citation7 All or some of these functions may depend upon a conformational change in the RLIP76 protein when it binds to Ral proteins, which could for example, lead to exposure of new binding sites for other molecules or unmasking of a membrane attachment site.
The role of RLIP76 as a plasma membrane pump is intriguing because it implies that the protein has two important properties: that it is a membrane protein and that it binds to ATP. The fact that full-length RLIP76 can be expressed solubly in E. coli both in our hands and in other laboratories (reviewed in ref. Citation4 and Citation6), suggests that it is not permanently attached to the membrane but rather that it would need to translocate there under certain circumstances. This would require a conformational change subsequent to translocation to expose cryptic transmembrane sequences. The other property, that of ATP binding, is more straightforward to envisage. Two ATP-binding sequences were identified in RLIP76:Citation22 the first, encompassing residues 69–74, contains a Walker A motif, while the second, which extends over residues 418–425, is homologous to the ATP-binding motif from phosphoglycerate kinase (PGK).Citation23 This second motif is within the RLIP76 RBD itself and has the sequence 418GGI KDLSK425. Furthermore, mutation of Lys-425 to Met abrogated ATP binding and the ATPase activity of RLIP76.Citation22 We have been unable to detect ATP binding directly to the small fragment that we have used in this work, but we have used the structure of porcine PGK in complex with Mg2+-ATP (pdb code 1VJC) to model how the RLIP76 RBD could contribute bind to ATP binding in the context of full-length RLIP76. Porcine PGK has the sequence 212GGAKVADK219 in its ATP-binding motif. We superimposed the Cα, C′, N and Cβ atoms of RLIP76 RBD and PGK over residues 418–425 and 212–219 respectively and the structures had a root mean squared deviation of 2.47 Å. The co-ordinates of ATP from the PGK structure were then modelled into our RLIP76 RBD structure. A comparison of PGK and the RBD () shows that the Lys residues are in similar secondary structures in both proteins, i.e., the N-terminal Lys is in a loop and the C-terminal Lys is in a helix. The sidechain of Lys-425 of RLIP76 is in a position where it could indeed interact with ATP () and is in a similar orientation to the equivalent Lys in the PGK structure. The conformation of the interhelical loop in the RLIP76 RBD is different to the conformation of the loop in PGK and the Lys residues in the loops are not pointing in the same direction. Our NMR data show that this loop is flexible in the free RLIP76 RBD and therefore it has the conformational freedom and can adopt multiple conformations, including an ATP binding conformation. The RalB binding site does not directly overlap with the ATP binding site but one of the conserved Lys residues, Lys-421, is in contact with RalB. It is also apparent from the structures that the conformational freedom of the loop between the two helices in the RLIP is restricted when it binds to RalB and this loss of mobility may well modulate the interaction with ATP. It is evident from the PGK structure that the simple coiled-coil that comprises the RBD will be insufficient to form an ATP binding site. The Lys residues in the RBD are adequate to engage the phosphate moieties of the ATP but other parts of the protein will also be required to bind the ribose and adenine ring to achieve high affinity binding and select ATP over other nucleotide triphosphates. The identity of these other regions has yet to be revealed but it is interesting to note that POB1 binding to RLIP76 inhibits its transporter activity,Citation24 so it is possible that POB1 somehow interferes with the ATP binding site.
Figures and Tables
Figure 1 The RLIP76-RalB structure. (A) The complex formed between RalB and the RLIP76 RBD involves contacts with both switch regions. RalB is shown in dark blue; RLIP76 is orange. The two switch regions and the N-terminus of each protein are indicated. The nucleotide is shown in a ball-and-stick representation and the atoms are colored as follows: carbon black; nitrogen blue; oxygen red; phosphorus pink. The Mg2+ ion is shown in a spacefilling representation. (B) Close-up of a comparison between the putative ATP binding region in the RLIP76 RBD and the ATP binding region in phosphoglycerate kinase (PGK). RLIP76 is shown in orange and PGK (pdb code 1VJC) is green. The α-helix and preceding loop are shown in dark orange and green and the rest of the protein is in pale shades of orange and green. The sidechains of the two Lys residues in the sequence and the ATP molecule are shown in a stick representation. Lys residues are colored orange or green and are labelled in their corresponding colors. ATP atoms are colored as in (A). Lys219 of PGK contacts the phosphates of the ATP molecule and Lys425 points in the same direction in the RLIP76 RBD. Lys215 in PGK is partially disordered but also points towards the ATP. The equivalent Lys in RLIP76, Lys421 is pointing in the other direction in this structure but is in a disordered loop and would therefore be able to change conformation. This residue is also involved in interacting with RalB.
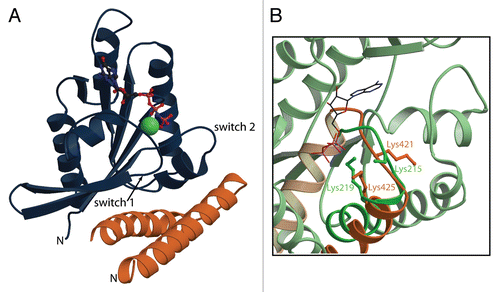
Figure 2 Alignment of RalA and RalB and their binding sites for effectors. The sequence alignment of human RalA and RalB with the divergent residues boxed. Residues that interact with the three effectors are colored as follows: Exo84 only-yellow; Sec5 only-red; RLIP76 only-blue; Exo84 and Sec5: orange; Exo84 and RLIP76-green; all three effectors-grey. It is evident that the residues that interact directly with the effectors are 100% conserved between the two Ral isoforms. The numbers and secondary structures above the sequences are for RalA. Helices are denoted by cylinders and β-strands by arrows. The switch regions, the P loop and the variable region at the C-terminus are also indicated above the sequences. This figure was produced with Alscript.
Acknowledgements
This work was supported by the Medical Research Council (G0700057 to H.R.M. and D.O.).
Extra View to: Fenwick RB, Campbell LJ, Rajasekar K, Prasannan S, Nietlispach D, Camonis J, Owen D, Mott HR. The RalB-RLIP76 complex reveals a novel mode of Ral-effector interaction. Structure 2010; 18:985 - 995; PMID: 20696399; http://dx.doi.org/10.1016/j.str.2010.05.013
References
- Cantor SB, Urano T, Feig LA. Identification and Characterization of Ral-Binding Protein-1, a Potential Downstream Target of Ral Gtpases. Mol Cell Biol 1995; 15:4578 - 4584
- Park SH, Weinberg RA. A putative effector of Ral has homology to Rho/Rac GTPase activating proteins. Oncogene 1995; 11:2349 - 2355
- Jullien-Flores V, Dorseuil O, Romero F, Letourneur F, Saragosti S, Berger R, et al. Bridging Ral Gtpase to Rho-Pathways—Rlip76, a Ral Effector With Cdc42/Rac Gtpase-Activating Protein Activity. J Biol Chem 1995; 270:22473 - 22477
- Jullien-Flores V, Mahe Y, Mirey G, Leprince C, Meunier-Bisceuil B, Sorkin A, et al. RLIP76, an effector of the GTPase Ral, interacts with the AP2 complex: involvement of the Ral pathway in receptor endocytosis. J Cell Sci 2000; 113:2837 - 2844
- Yamaguchi A, Urano T, Goi T, Feig LA. An eps homology (EH) domain protein that binds to the Ral-GTPase target, RalBP1. J Biol Chem 1997; 272:31230 - 31234
- Ikeda M, Ishida O, Hinoi T, Kishida S, Kikuchi A. Identification and characterization of a novel protein interacting with Ral-binding protein 1, a putative effector protein of Ral. J Biol Chem 1998; 273:814 - 821
- Coon BG, Burgner J, Camonis JH, Aguilar RC. The epsin family of endocytic adaptors promotes fibrosarcoma migration and invasion. J Biol Chem 2010; 285:33073 - 33081
- Smith SC, Oxford G, Baras AS, Owens C, Havaleshko D, Brautigan DL, et al. Expression of Ral GTPases, their effectors and activators in human bladder cancer. Clin Cancer Res 2007; 13:3803 - 3813
- Hudson ME, Pozdnyakova I, Haines K, Mor G, Snyder M. Identification of differentially expressed proteins in ovarian cancer using high-density protein microarrays. Proc Natl Acad Sci USA 2007; 104:17494 - 17499
- Vatsyayan R, Lelsani PCR, Awasthi S, Singhal SS. RLIP76: A versatile transporter and an emerging target for cancer therapy. Biochem Pharmacol 2010; 79:1699 - 1705
- Han K, Kim MH, Seeburg D, Seo J, Verpelli C, Han S, et al. Regulated RalBP1 binding to RalA and PSD-95 controls AMPA receptor endocytosis and LTD 2009; 7:1000187
- Modha R, Campbell LJ, Nietlispach D, Buhecha HR, Owen D, Mott HR. The Rac1 Polybasic Region Is Required for Interaction with Its Effector PRK1. J Biol Chem 2008; 283:1492 - 1500
- Vetter IR, Wittinghofer A. Signal transduction—The guanine nucleotide-binding switch in three dimensions. Science 2001; 294:1299 - 1304
- Nassar M, Horn G, Herrmann C, Scherer A, McCormick F, Wittinghofer A. The 2.2-Angstrom Crystal-Structure of the Ras-Binding Domain of the Serine Threonine Kinase C-Raf1 in Complex With Rap1a and a Gtp Analog. Nature 1995; 375:554 - 560
- Fukai S, Matern HT, Jagath JR, Scheller RH, Brunger AT. Structural basis of the interaction between RalA and Sec5, a subunit of the sec6/8 complex. EMBO J 2003; 22:3267 - 3278
- Bodemann BO, White MA. Ral GTPases and cancer: linchpin support of the tumorigenic platform. Nat Rev Cancer 2008; 8:133 - 140
- Chien Y, White MA. RAL GTPases are linchpin modulators of human tumour-cell proliferation and survival. EMBO Rep 2003; 4:800 - 806
- Fenwick RB, Prasannan S, Campbell LJ, Nietlispach D, Evetts KA, Camonis J, et al. Solution Structure and Dynamics of the Small GTPase RalB in Its Active Conformation: Significance for Effector Protein Binding. Biochemistry 2009; 48:2192 - 2206
- Fenwick RB, Campbell LJ, Rajasekar K, Prasannan S, Nietlispach D, Camonis J, et al. The RaIB-RLIP76 Complex Reveals a Novel Mode of RaI-Effector Interaction. Structure 2010; 18:985 - 995
- Lim KH, Brady DC, Kashatus DF, Ancrile BB, Der CJ, Cox AD, et al. Aurora-A Phosphorylates, Activates and Relocalizes the Small GTPase RalA. Mol Cell Biol 2010; 30:508 - 523
- Nakashima S, Morinaka K, Koyama S, Ikeda M, Kishida M, Okawa K, et al. Small G protein Ral and its downstream molecules regulate endocytosis of EGF and insulin receptors. EMBO J 1999; 18:3629 - 3642
- Awasthi S, Cheng JZ, Singhal SS, Pandya U, Sharma R, Singh SV, et al. Functional reassembly of ATP-dependent xenobiotic transport by the N- and C-terminal domains of RLIP76 and identification of ATP binding sequences. Biochemistry 2001; 40:4159 - 4168
- Saraste M, Sibbald PR, Wittinghofer A. The P-Loop— a Common Motif in Atp-Binding and Gtp-Binding Proteins. Trends in Bioch Sci 1990; 15:430 - 434
- Yadav S, Zajac E, Singhal SS, Singhal J, Drake K, Awasthi YC, et al. POB1 overexpression inhibits RLIP76-mediated transport of glutathione-conjugates, drugs and promotes apoptosis. Biochem Biophys Res Commun 2005; 328:1003 - 1009