Abstract
RhoGDIs are negative regulators of small GTP-binding proteins of the Rho family, which have essential cellular functions in most aspects of actin-based morphology and motility processes. They extract Rho proteins from membranes, keep them in inactive rhoGDI/Rho complexes, and eventually deliver them again to specific membranes in response to cellular signals. RhoGDI3, the most divergent member of the rhoGDI family, is well suited to document the underlying molecular mechanisms, since the active and inactive forms of its cellular target, RhoG, have well-separated subcellular localizations. In this study, we investigate trafficking structures and molecular interactions involved in rhoGDI3-mediated shuttling of RhoG between the Golgi and the plasma membrane. Bimolecular fluorescence complementation and acceptor-photobleaching FRET experiments suggest that rhoGDI3 and RhoG form complexes on Golgi and vesicular structures in mammalian cells. 4D-videomicroscopy confirms this localization, and show that RhoG/rhoGDI3-labelled structures are less dynamic than RhoG and rhoGDI3-labeled vesicles, consistent with the inhibitory function of rhoGDI3. Next, we identify the Exocyst subunit Sec3 as a candidate rhoGDI3 partner in cells. RhoGDI3 relocates a subcomplex of the Exocyst (Sec3 and Sec8) from the cytoplasm to the Golgi, while Sec6 is unaffected. Remarkably, Sec3 increases the level of GTP-bound endogenous RhoG, the RhoG-dependent induction of membrane ruffles, and the formation of intercellular tunneling nanotube-like protrusions. Altogether, our study identifies a novel link between vesicular traffic and the regulation of Rho proteins by rhoGDIs. It also suggests that components of the Exocyst machinery may be involved in RhoG functions, possibly regulated by rhoGDI3.
Introduction
Members of the Rho/Rac/Cdc42 family of small GTP-binding proteins function as molecular switches to control formation of actin stress fibers, lamellipodia and filopodia, thus modulating cell morphology, cell motility and membrane trafficking events (reviewed in refs. Citation1 and Citation2). These functions are involved in cancer progression, including proliferation, invasion and metastasis, in which the contribution of Rho proteins is well established (reviewed in ref. Citation3–Citation5). The functional cycle of Rho family members combines a GDP/GTP cycle, which is stimulated by guanine nucleotide exchange factors (rhoGEFs) and terminated by GTPase-activating proteins (rhoGAPs), and changes in subcellular localization, regulated by guanine nucleotide dissociation inhibitors (rhoGDIs). RhoGDIs regulate the activity of Rho proteins by a shuttling process involving the extraction of Rho from donor membranes, the formation of inhibitory cytosolic Rho/rhoGDI complexes and the delivery of Rho to membrane target sites (reviewed in ref. Citation6–Citation8). Three rhoGDIs have been identified so far in mammals and plants. In mammals, rhoGDI-1 (or rhoGDIα) is the most abundant and ubiquitous representative, and is able to form cytosolic complexes with most members of the Rho family. In contrast, D4/LYGDI (also called rhoGDI-2 or rhoGDIβ) is found predominantly in hematopoietic cells but also in some non-hematopoietic neoplasm,Citation9–Citation11 and has a more narrow specificity for the Rac subfamily. We identified and characterized a third rhoGDI, rhoGDI3 (also called rhoGDIγ),Citation12 shown to regulate specifically the activity of the small G-protein RhoG.Citation13,Citation14 RhoGDI3 (referred to as GDI3 hereafter) carries a unique N-terminal extension that targets it to Golgi membranes, while most rhoGDIs are cytosolic.Citation12 The molecular target at the Golgi apparatus for the N-terminal extension of GDI3 is currently unknown.Citation13 We reported previously that ectopically expressed GDI3 co-localizes with Golgi markers and that it is sensitive to Golgi disassembly by BFA.Citation13 Interestingly, an unbalanced expression between RhoG (increased) and GDI3 (decreased) was shown to correlate with node-positive breast cancer tumors and metastasis.Citation15
The major GDI3 target, RhoG, is activated at the perinuclear region by the exchange factor Trio, then it translocates to active sites at the plasma membrane in a microtubule-dependent manner where it induces several morphological processes.Citation16,Citation17 GDI3 relocates inactive RhoG from the plasma membrane to the Golgi.Citation13 Consistently, RhoG was reported to move sequentially from the plasma membrane to intracellular vesicles and the Golgi apparatus along markers of caveolar endocytic pathway.Citation18 Activation of RhoG by Trio induces a direct interaction of RhoG-GTP with Elmo, the formation of a ternary complex comprising RhoG-GTP, Elmo and Dock180, and the translocation of the Elmo-Dock180 complex to the plasma membrane.Citation19 This complex functions as a Rac1-specific GEFCitation20,Citation21 which promotes several cellular morphological actions, such as neurite outgrowth, dorsal protrusions, peripheral lamellipodia, membrane ruffles and cell migration.Citation16,Citation22–Citation24 As they induce actin rearrangement at the dorsal membrane,Citation25,Citation26 RhoG and Elmo-Dock participate additionally in macropinocytosis,Citation27 phagocytosis,Citation28,Citation29 and in endothelial apical cup assembly.Citation30 In addition, guanine-nucleotide exchange factors of the Vav family were shown to mediate EGF-induced rapid activation of RhoG.Citation31
Several mechanisms have been proposed to regulate rhoGDI activity and functional specificity in cells, including phosphorylation,Citation32,Citation33 interaction with signaling complexes,Citation34 coordination with rhoGEFs,Citation35,Citation36 and assistance by the membrane.Citation37,Citation38 However, the mechanism and dynamics of Rho shuttling by rhoGDIs in cells, including extraction, transport and delivery events, remain poorly understood. GDI3 provides a convenient system to address these issues, since it shuttles RhoG between different locations in cells and has a unique Golgi targeting mediated by its N-terminal helix.Citation12,Citation13 We previously devised non-inhibitory GDI3 mutants, which retain their ability to interact with RhoG but fail to extract it from membranes.Citation14 The use of these non-inhibitory GDI3 mutants identified a co-localization of GDI3 and RhoG not only at Golgi and vesicular-like structures in the cytoplasm, but also at the plasma membrane suggesting an active involvement in intracellular trafficking events.Citation14 Trafficking of RhoG between the Golgi apparatus and the plasma membrane requires an effective microtubule network,Citation17 suggesting that the RhoG/GDI3 system may follow a microtubule route to the plasma membrane on carriers of yet unknown nature. To gain further insight into these shuttling processes between Golgi and the plasma membrane, we analyze in this work the dynamics of intracellular compartments defined by fluorescently labeled GDI3 and RhoG proteins in live MDCK cells. Furthermore, we identify subunits of the Exocyst complex as novel partners in the GDI3 regulated RhoG trafficking pathway and analyze the involvement of the Sec3 Exocyst subunit in the regulation of endogenous RhoG functions.
Results
Interaction and dynamics of GDI3 and RhoG at the Golgi apparatus and on vesicles.
The action of GDI3 on RhoG activity and intra-cellular localization was previously investigated in HeLa cells.Citation13,Citation14 To further analyze the interaction and dynamics of these two proteins, we selected epithelial MDCK cells. In these cells, the fluorescent GDI3-EYFP fusion protein localized to Golgi and on punctuate vesicular-like structures in the cytoplasm (), as found in HeLa cells for GDI3 fused either to a Myc epitope or to the EYFP fluorescent protein.Citation13,Citation14 ECFP-RhoG, expressed in MDCK cells, localizes to perinuclear and vesicular-like structures and to membrane ruffles at the plasma membrane (arrows in ) as already described for HA-RhoG in HeLa cells. RhoG also distributes in a tubulovesicular pattern throughout the cytoplasm, highlighted in as previously described in REF-52 fibroblasts, in COS-7 and in HeLa cells.Citation14,Citation16,Citation17 Finally, as found for HeLa cells, co-expression of GDI3-EYFP with ECFP-RhoG resulted in extensive co-localization in the Golgi region and on vesicular-like structures throughout the cytoplasm with an associated disappearance of membrane ruffles and RhoG from the cell periphery ( and D).
To investigate whether co-localized GDI3 and RhoG proteins interact directly at these locations we used the Bimolecular Fluorescence Complementation (BiFC) assay. This assay allows the direct visualization of protein interactions in their cellular environment with minimal perturbation.Citation39 We co-transfected MDCK cells with equal amounts of plasmids coding for RhoG fused to the N-terminal half of the fluorescent protein YFP (YFPNt) and GDI3 fused to the C-terminal half of YFP (YFPCt). Co-expression of RhoG fused with YFPNt (non fluorescent) and GDI3 fused with YFPCt (non fluorescent) allowed the reconstitution of the YFP fluorescence (in yellow), suggesting that RhoG and GDI3 interact directly. As shown by the two images in , RhoG/GDI3 interactions could be identified, by this technique, at both Golgi and punctuate structures in the cytoplasm. 100 cells where examined from each of five independent experiments. 100% of these cells showed YFP fluorescence at the Golgi structures and 75% at vesicles. Similar results were obtained using HeLa cells (not shown). To corroborate this potential RhoG/GDI3 interaction at the Golgi apparatus we next analyzed FRET from ECFP-RhoG (cyan) to GDI3-EYFP (yellow) fluorescent proteins in MDCK cells by the acceptor photobleaching technique and confocal microscopy. In this method, the occurrence of FRET between donor and acceptor is revealed upon photo-bleaching destruction of the acceptor fluorescence. If FRET is occurring, then photobleaching of the acceptor yields a significant increase in fluorescence of the donor.Citation40 The resulting FRET efficiency is then defined as the percentage of donor fluorescence increase. We bleached cells in the YFP channel by scanning several regions of interest (ROI) at the Golgi level, corresponding to either the entire Golgi area or to sub-regions of the Golgi. As a negative control we analyzed the FRET efficiency between ECFP-Rab6 and GDI3-EYFP. Rab6, a small GTP-binding protein, partially co-localizes with GDI3 at the Golgi apparatus but does not interact with GDI3.Citation13 Representative results of FRET at the Golgi apparatus in cells co-expressing GDI3-EYFP with either ECFP-RhoG or ECFP-Rab6 are shown in and G. The RhoG/GDI3 FRET efficiency was 19.01 ± 4.03%, as measured in 19 cells corresponding to a total of 52 ROI (42 subregions and 10 whole Golgi). In contrast, Rab6/GDI3 FRET efficiency was 1.3 ± 1.2% as measured in 7 cells corresponding to 38 individual ROI, indicating that no FRET is occurring. This ensemble of experiments suggests that GDI3 and RhoG interact not only as soluble complexes in the cytoplasm,Citation13 but also at the Golgi region and on vesicular-like structures, which could correspond to trafficking vesicles. It also shows that this interaction is probably direct. To our knowledge, this is the first time that Rho/rhoGDI interactions are identified at endomembranes.
Our observation that GDI3 and RhoG interact on Golgi and vesicular structures, together with their co-localization on plasma membrane structures, previously observed using our non-inhibitory GDI3 mutants,Citation14 suggests that they are actively involved in intracellular trafficking events. This prompted us to analyze their dynamics in live MDCK cells co-expressing GDI3-mCherry and EGFP-RhoG. We used rapid 3D + time (4D) microscopy followed by deconvolution. As suggested by observation in fixed cells using confocal microscopy, analysis of the video frames of living cells confirms that GDI3 and RhoG are present on vesicular structures (). GDI3 vesicles (red) appear scattered throughout the cells, while RhoG-labeled structures (green) are mostly found close to the plasma membrane. Structures that are double-labeled with GDI3 and RhoG (yellow) appear to be larger, and mainly concentrated in the perinuclear area and beneath the plasma membrane.
Two cells were chosen for a qualitative analysis of the movements of these structures (Videos 1 and 2 in Sup. data). A third cell was chosen to show the more static co-localization at Golgi structures (Video 3 in Sup. data). For raw green and red projections, masks were generated in order to isolate either the RhoG signal alone (), the GDI3 signal alone (), or the locations where the two signals co-localize (). RhoG traces are found inside the cell, where they are on average shorter than those of GDI3. They also highlight the plasma membrane (arrow in ), suggesting diffusional movements on this membrane. GDI3 highlights radial and extended traces, probably centered on the microtubule-organizing center and following microtubules (arrow in ). Double-labeled structures leave traces in a centrosomal-like location, and closer to the plasma membrane (arrows in ). These traces are large and punctuate, probably reflecting small local movements. shows overlay of the 3 pseudo-channels.
In order to gain qualitative insight into individual trajectories, kymographs were generated from traces extracted from 100-second projections, as shown in . The trajectories for a total of 57, 16 and 25 individual structures labeled respectively with RhoG alone, GDI3 alone or both proteins were analyzed for their directions and speed. The distribution of speeds is shown in . GDI3 and RhoG-labeled vesicles were found to be highly dynamic on average (see temporal traces in and in Sup. material, Videos 1–3). Alternation of inwards and outwards movements, interrupted by pauses, is seen for both populations (for example kymographs 1 and 4 in ). In contrast, structures labeled with both proteins appeared significantly more static (see for example kymograph 6 in ). Altogether these video-microscopy experiments show that GDI3 and RhoG are found on mobile vesicles, and that their co-localization coincides with a decrease in their dynamics, suggesting that the GDI3/RhoG complex, but not GDI3 alone, is involved in slowing vesicular motions.
Interaction of GDI 3 with components of the Exocyst complex.
The unique N-terminus of GDI3, encompassing a predicted alpha helix,Citation13 is necessary for Golgi targeting of GDI3 or the rhoG/GDI3 complex and sufficient to address a fused EYFP fluorescent protein to Golgi.Citation13 This extension may therefore be engaged in protein interactions that are critical for the dynamics of GDI3-dependent RhoG trafficking in eukaryotic cells. To test this hypothesis, we first assessed whether this extension defines a saturable site at the Golgi. For this purpose we analyzed the outcome of a competition at Golgi sites between full-length GDI3 and its unique N-terminal peptide (amino acids 1–41) fused to EYFP, named from now NtGDI3-EYFP (α-EYFP in ref. Citation13). shows the expression of NtGDI3-EYFP (green artificial color) in MDCK cells compared to that of full-length GDI3 fused to the fluorescent protein mCherry. When MDCK cells were co-transfected with a ratio of NtGDI3-EYFP/GDI3-mCherry coding plasmids of 2, GDI3-mCherry was not only localized to Golgi, as NtGDI3-EYFP, but also easily identified at the cell periphery (arrows in ), where it is normally not observed in reference Citation13 and Citation14. As seen in , GDI3-mCherry was also identified at tubulovesicular structures in the cytoplasm besides its localization at Golgi and near the plasma membrane. The images shown in and b are representative of 68% of 100 co-expressing cells. In the remaining cells, NtGDI3-EYFP was only faintly detected suggesting low levels of expression compared to GDI-mCherry. Accordingly, a reduced amount of GDI3 was displaced from the perinuclear region in these cells. Finally, simultaneous co-expression of RhoG, full-length GDI3 and NtGDI3-EYFP resulted in a co-localization of GDI3 and RhoG at the cell periphery (arrows in and b) in addition to their localization to Golgi and a tubulovesicular distribution in the cytoplasm. This was seen in 57% of 100 cells co-expressing the three proteins. shows that, in the absence of co-expressed NtGDI3-EYFP, the GDI3-mCherry protein relocates ECFP-RhoG (green artificial color) to Golgi and vesicles, with a concomitant disappearance of RhoG from the cell periphery and from the tubulovesicular cytoplasmic pattern as described for HeLaCitation13 and MDCK cells ( and D) using different tags. These observations suggest that the N-terminal extension competes with GDI3 for a protein-protein interaction at the Golgi.
In order to identify candidate GDI3 partners, we therefore chose this peptide as a bait to screen a yeast expression library derived from human brain using the yeast two-hybrid system. Since we used the mouse GDI3 peptide in this assay, positive clones were further assayed against full-length mouse and human GDI3 as well as the NtGDI3 peptide in a yeast-mating assay ( and Sup. material Fig. 1). Two clones (Act-4 and Act-37), which interacted with all constructs, corresponded to the 390 and 261 C-terminal residues of the human Sec3 protein (BAA91886), a subunit of the Exocyst complex.Citation41 To investigate the existence of an interaction between GDI3 and Sec3 in mammalian cells, we then expressed Flag-Sec3 and GDI3-HA in HeLa cells and co-immunoprecipitated them from subcellular fractions using either HA or Flag antibody-coated beads. Sec3 and GDI3 were detected in the immunoprecipitates in both cases (). We further used the BiFC technique to analyze the GDI3/Sec3 interaction in MDCK cells. Yellow fluorescence was recovered at the Golgi apparatus by co-expression of YFPNt-Sec3 and GDI3-YFPCt (), which suggests a direct interaction between the two proteins. The image is representative of 100% of the 40 cells analyzed by this technique in 3 independent experiments. We next examined whether this GDI3/Sec3 interaction may operate in GDI3 and/or Sec3 localization. First, we analyzed the effect of GDI3 on the distribution of Sec3 in MDCK cells. In the absence of transfected GDI3, ECFP-Sec3 was distributed diffusely throughout the cytoplasm ( and b), as previously reported in reference Citation41. When co-expressed with GDI3-EYFP, ECFP-Sec3 was displaced to the Golgi and to vesicular structures, where it co-localized with GDI3-EYFP (). This image is representative of 100% of 150 co-expressing MDCK cells. Co-expression of NtGDI3-EYFP with ECFP-Sec3 was sufficient to translocate Sec3 to these endomembranes ( and b), in agreement with the interaction of the N-terminus of GDI3 with Sec3 identified in the yeast-two hybrid experiment (). This translocation is seen in 100% of the 75 co-expressing cells. Similar results were obtained in HeLa cells (not shown). This ensemble of results establishes that GDI3 interacts with Sec3, and that this interaction involves the N-terminal extension of GDI3 and the C-terminal region of Sec3.
Sec3 is part of the octameric Exocyst complex (reviewed in ref. Citation42 and Citation43). Several observations, made in yeast and mammals, suggest however that not all subunits may be present at the same time in the complex.Citation44–Citation47 We thus examined whether GDI3 could also affect the localization of other Exocyst subunits than Sec3. We focused on Sec8, reported to interact directly with Sec3,Citation48 and Sec6, which can form a sub-complex with Sec8.Citation43 The localization of endogenous Sec8 and Sec6 in MDCK cells has been reported to be highly dependent on the level of MDCK polarization and on experimental conditions.Citation46,Citation47 We therefore performed our experiments under invariable conditions using non-confluent monolayer of non-polarized MDCK cells cultured at low density, where few or early cell-to-cell contacts are seen before fixation. Under these conditions and with the antibodies to Sec8 and Sec6 used here, immuno-reactive endogenous Sec8 has a diffuse cytoplasmic localization (). Expression of ECFP-Sec3 did not modify the localization of endogenous Sec8, which remained essentially cytoplasmic and showed a diffuse co-localization with Sec3 (). In contrast, expression of GDI3-EYFP (in green artificial color) targets the endogenous Sec8 subunit to Golgi and vesicles (), as it does to ectopically expressed ECFP-Sec3 (). This recruitment of Sec8 to Golgi, seen in 100% of the 95 co-expressing cells, could be due to a direct GDI3/Sec8 protein-protein interaction or, most probably, to an indirect interaction involving endogenous Sec3 as a link between GDI3 and Sec8, since Sec3 and Sec8 interact with each other.Citation48 Consistently, co-expression of GDI3-EYFP and ECFP-Sec3 also displaced Sec8 from the cytoplasm to Golgi, where the three proteins co-localized (). In contrast to Sec8, the endogenous Sec6 subunit was identified at the plasma membrane, mostly at cell-cell contacts and as punctuate structures in the cytoplasm (). Remarkably, the expression of GDI3-EYFP did not displace endogenous Sec6 from the plasma membrane in more than 100 analyzed cells (). Co-expression of GDI3-EYFP and ECFP-Sec3 was also ineffective at displacing Sec6 (). Taken together, this set of observations may imply that Sec3 acts, in GDI3-dependent pathways, as part of an Exocyst sub-complex containing Sec8 but not Sec6.
Sec3 is involved in RhoG-regulated plasma membrane morphology.
The finding that Sec3 interacts functionally and physically with GDI3, a major regulator of RhoG, raises the interesting possibility that it may be involved in RhoG functions. To address this issue, analysis were conducted by confocal microscopy studies of MDCK cells co-expressing RhoG and Sec3. and b shows membrane ruffles induced by EGFP-RhoG, similar to those observed in MDCK cells expressing ECFP-RhoG () and in HeLa cells using HA-tagged or GFP-tagged RhoG.Citation13,Citation14 Remarkably, MDCK cells co-expressing EGFP-RhoG and ECFP-Sec3 develop long, thin membrane protrusions extending towards neighboring cells (arrows in ). These protrusions were detected in 72% of the 182 co-transfected cells. They seem to be similar to tunneling nanotubes (TNTs) as several of them made contacts with neighboring cells.Citation49–Citation51 RhoG and Sec3 co-localized inside these TNT-like protrusions, at perinuclear regions, and at tubulovesicular cytoplasmic structures. The known role of RhoG in promoting cortical actin structures such as those found in these protrusions is consistent with our observations. Altogether, these observations suggest that Sec3 is involved in RhoG functions at the cell periphery.
To test whether Sec3 regulates the activity of RhoG, we performed GST-pulldown assays using GST-ELMO, which is an effector for endogenous RhoG, in MDCK cells transfected by Flag-Sec3 ( and lane 2). As a control we used subcellular fractions from cells transfected by GDI3 and from non-transfected cells ( and lane 1 and 3, respectively). In each case, we measured the amount of GTP-bound active RhoG relative to total endogenous RhoG. We found a large 15-fold increase in the relative levels of active RhoG in Sec3-expressing cells (lane 2) compared with non-transfected cells (lane 3). The level of active endogenous RhoG could hardly be detected in cells expressing GDI3-Myc (lane 1), consistent with the inhibitory activity of GDI3 towards RhoG.
Finally, we investigated whether GDI3 retains its inhibitory functions towards RhoG in the presence of Sec3. We performed triple-transfection of MDCK cells with plasmids coding for EGFP-RhoG, ECFP-Sec3 and GDI3-mCherry. Notably, GDI3 relocated both Sec3 and RhoG to Golgi, with a concomitant disappearance of RhoG-induced membrane protrusions and nanotubes-like structures in 100% of the 80 co-transfected cells (). Thus GDI3 appears to downregulate RhoG regardless of the potentiating activity of Sec3.
Since GDI3 is able to displace Sec8 to the Golgi, we then investigated whether it also retains its ability to displace endogenous Sec8 to the Golgi in the presence of overexpressed RhoG using MDCK cells expressing ECFP-RhoG. Interestingly, expression of ECFP-RhoG alone induced a relocation of endogenous Sec8 to the Golgi region and to the cell periphery where the two proteins co-localize (arrows in ). This translocation of Sec8 was identified in 90% of the 70 ECFP-RhoG-expressing cells. As shown in , co-expression of GDI3-EYFP with ECFP-RhoG targets RhoG and endogenous Sec8 to Golgi with a concomitant disappearance of both proteins from the cell periphery. In contrast, co-expressed GDI3/RhoG remained ineffective at displacing the endogenous Sec6 Exocyst subunit from the plasma membrane ().
This ensemble of results suggests that Sec3 potentiates RhoG-mediated plasma membrane structures, and that the ability of GDI3 to displace RhoG from the plasma membrane and to inhibit its activity prevails on the stimulating activity of the Sec3 subunit.
Discussion
The trafficking events and the regulatory signals that control how rhoGDIs extract Rho proteins from membranes in the form of inactive complexes and deliver them to their sites of actions are still poorly understood. In this study, we analyze the cellular localization, dynamics and protein interactions of GDI3, which is a resourceful regulator for the investigation of these issues as it shuttles its substrate, RhoG, between well-separated sub-cellular localizations. We identify and characterize the dynamics of vesicles probably associated with the microtubule network and labeled respectively with GDI3 alone, RhoG alone or both proteins. We show, in particular, that GDI3 affects the dynamics of RhoG-positive vesicles, as vesicles labeled with both proteins appeared significantly more static. These observations suggest that, unlike usually assumed for rhoGDI regulators, shuttling of the GDI3/RhoG complex may follow a vesicular route rather than, or in parallel to, cytoplasmic diffusion of a solubleCitation13 rhoGDI/Rho complex. The movements along microtubules would be consistent with previous observations that microtubules are necessary for RhoG functions.Citation17 The fact that RhoG as well as GDI3 are also seen alone on these structures suggests however that RhoG could also be downregulated by other pathways, such as the one recently reported for the rhoGDI1/syndecan 4/synectin ternary complex.Citation34
The unique N-terminal extension of GDI3, which encompasses a predicted α-helix, is necessary for its localization at the Golgi,Citation13 and is thus also expected to be pivotal for the downregulation and/or trafficking of RhoG inside the cell. However, the nature of the interactions that support its specificity is not known. In this work, we show for the first time that this N-terminal extension is involved in protein-protein interactions, and identify the Sec3 subunit of the Exocyst as a cellular partner of GDI3 using the yeast two-hybrid screen. We demonstrate, by a yeast-mating assay, that this N-terminal extension interacts with a predicted C-terminal coiled-coil domain in Sec3. In MDCK cells, GDI3 translocates both Sec3 and Sec8 Exocyst subunits from a diffuse cytoplasmic localization to the Golgi. These results, and the previously described interaction of Sec8 with a predicted N-terminal coiled-coil domain of Sec3,Citation48 suggest a possible organization of a trimeric complex containing Sec8, Sec3 and GDI3. GDI3 translocates also RhoG and Sec8 from their co-localization at the cell periphery to the Golgi, suggesting the existence of large multi-protein signaling complexes consisting of Sec8, Sec3 and the GDI3/RhoG complex interacting with Sec3 through the N-terminal extension of GDI3. In addition, GDI3 seems to interact with a subcomplex of the Exocyst that does not contain the Sec6 subunit, as Sec6 localization is unaffected by GDI3 or by GDI3 complexes containing either RhoG or Sec3.
The Exocyst is a multi-subunit assembly that serves in tethering exocytic vesicles to destination membranes, and has been associated to polarized exocytosis. A common theme between the yeast and mammalian Exocyst complex is their interactions with small GTP-binding proteins (reviewed in ref. Citation52 and Citation53). Notably, yeast Sec3 interacts with Rho and Cdc42 by its N-terminal region, which also binds phosphoinositides.Citation54,Citation55 However, the corresponding region does not exist in mammalian Sec3, although a link between Sec3 and Rho pathways is suggested by its interaction with IQGAP1, a Cdc42 and RhoA effector protein.Citation48 Our findings that GDI3 interacts with a subcomplex of the Exocyst in cells, together with the increased RhoG activity upon Sec3/RhoG co-expression, opens the possibility that the pathway involving GDI3 and RhoG may be one of the mammalian pathways replacing the Sec3/Rho pathway in yeast. A link with RhoG would be consistent with the recent recognition of the role of the Exocyst in the migration of mammalian cells,Citation56–Citation58 a process that also involves RhoG.Citation23 The increase in RhoG activity upon Sec3 overexpression may then be due to either a titration of GDI3 by Sec3, thus decreasing its inhibitory function or reflect a more direct link between RhoG and Sec3 in the formation of motile actin structures. Elucidation of the molecular links between GDI3, RhoG and the Exocyst in cell migration will, however, require future investigations.
Materials and Methods
Materials.
Mouse monoclonal antibodies against Myc, HA and FLAG tags are from Sigma (M5546 anti-MYC clone 9E10, H9658 anti-HA clone HA-7, F1804 anti-FLAG clone M2), polyclonal HA-Tag antibody from Clontech Laboratories Inc., (630459), secondary antibodies against mouse or rabbit IgGs linked to horseradish peroxidase are from Amersham Pharmacia Biotech (NA934, NA931 respectively), monoclonal mouse anti-Sec8 antibody from BD Transduction Laboratories (Clone 14, 610659), monoclonal mouse anti-rat Sec6 antibody from Calbiochem (Calbiochem, 559240), the monoclonal antibody anti-RhoG is from Millipore (clone 1F3B3E5), the secondary anti-mouse immuno-reagents Alexa Fluor-568 is from Molecular Probes (A10037), Dynabeads M-280 goat anti-mouse IgG from Dynal-Invitrogen, (112.01D). Restriction endonucleases for molecular biology were from New England Biolabs. PCR reactions were performed using the Pfu Turbo polymerase, nucleotides mix and buffers from Stratagene.
DNA constructs.
For expression in eukaryotic cells, GDI3 cDNA was cloned into HA-tag containing pCD3FT plasmid. The GDI3-EYFP and GDI3Nt-EYFP were described in reference Citation13. RhoGDI3-mCherry was cloned in the pmCherry-N1 expression plasmid, a kind gift from Marie-José Masse (Maïté Coppey, IJM, Université-Paris-Diderot). The EGFP-RhoG construct is a kind gift from Anne Debant (CRBM, CNRS). The ECFP-RhoG and ECFP-Rab6 chimeras were cloned in the pECFP-C1 vector (Clontech Laboratories Inc., 6901-1). The full-length human Sec3 cDNA clone (Acc. n° AK001755) was purchased [NITE, Biological Resource Center (NBRC)]. Flag-Sec3 and ECFP-Sec3 were cloned respectively into pMZS.3Flag and ECFP plasmids. pMZS.3Flag is a kind gift from Mahel Zeghouf (LEBS, CNRS). The pGEX-GST-ELMO1 plasmid is a kind gift from Professor Kodi Ravichandran (UVA Cancer Center, University of Virginia). The two-hybrid vectors pAct-2, RalB-pFBL23 and RalBD-pGAD1318 are kind gifts from Jacques Camonis (Institut Curie, Centre de Recherche, INSERM U528).
Cell culture and transfection.
HeLa or MDCK cells were grown on 100 mm tissue dishes in DMEM containing 10% fetal calf serum and antibiotic/antimycotic mixture (Gibco BRL, 15240-096) as described in reference Citation13. Cells were transfected by electroporation as previously explained in reference Citation59. For fluorescence studies, transfected cells were plated in individual wells on Labtek multichamber glass slides (Thermo Fisher Scientific Inc., Nunc Lab-Tek, 177402). Non-confluent MDCK cells for Sec8 and Sec6 localization were cultured at low density and fixed with PFA 3% before 0.25% saponin treatment.
Confocal microscopy.
In all confocal investigations, cells were fixed within 20 h of transfections for 10 min with 3% paraformaldehyde in PBS at room temperature. Cells expressing fluorescent fusion proteins were washed in PBS and directly mounted in Vectashield medium (Vector Laboratories, H-1000). Fixed cells expressing non-fluorescent proteins were glycinated and permeabilized with 0.25% saponin for 5 min followed by PBS washes. Slides were preblocked with 5% BSA, 0.05% Tween 20 and 0.01% saponin in PBS. Incubations with mouse monoclonal anti-Sec6 antibody or anti-Sec8 antibody (1:200) were performed in PBS containing 0.25% bovine serum albumin, 0.01% Tween and 0.01% saponin overnight at 4°C. Staining with secondary antibody was carried out for 20 min at room temperature using Alexa Fluor 568 anti-mouse IgG (1:800). Confocal laser-scanning microscopy was performed using a Leica TCS SP2 confocal microscope using objective HCX Plan Apo CS 63x numerical aperture 1.40. For all co-expression studies involving multi-channel imaging, the different fluorophores were captured sequentially.
Bimolecular fluorescence complementation (BiFC) experiments.
For the BiFC analysis we linked the C-terminus of EYFP (residues 158–238) to the C-terminus of GDI3 (GDI3-YFPCt). To obtain the YFPNT-RhoG construct, the N-terminus of EYFP (residues 1–173) were linked to the N-terminus of RhoG. The YFPNt-Sec3 plasmid was obtained by linking the same N-terminal EYFP fragment to the N-terminus of Sec3. Finally, to provide flexibility for independent motion of the EYFP fragments and the interacting proteins we connected the GDI3, RhoG or Sec3 proteins to the EYFP fragments using the linkers recommended by several authors.Citation60,Citation61 None of the constructs were fluorescent when expressed alone as fusion proteins in MDCK cells.
Acceptor photobleaching FRET assay.
FRET from cyan (ECFP-RhoG) to yellow (GDI3-EYFP) fluorescent proteins was studied by the acceptor photobleaching technique and confocal microscopy using a single argon laser. We bleached cells in the YFP channel by scanning a ROI (region of interest) 15 to 10 times using the 514 nm argon laser line at 100% intensity. Depending on the experiment, we selected two to nine ROIs at the Golgi region for bleaching in each cell. Before and after this bleach, CFP and YFP images were collected by sequential acquisition at low intensity beginning with the CFP 458 nm laser channel. Cells carrying ECFP-RhoG alone were used as negative controls. The FRET efficiency was calculated as FRETeff = (Dpost − Dpre)/Dpost where Dpre and Dpost are the total fluorescence of the ROI before and after bleaching. ROIs were randomly selected in the Golgi region of each cell.
Rapid 3D and time (4D) microscopy in live MDCK cells.
For live cell double-labelling experiments, MDCK cells were co-transfected with equal amounts (20 µg each) of GDI3-mCherry and EGFP-RhoG plasmids. Co-transfected cells were imaged after 18–20 h incubation at 37°C (5% CO2). Transfected MDCK live cells were recorded using a Photometrics cool-SNAPHQ2 CCD camera connected to a Leica DM IRE2 inverted microscope. Stacks of 5 images were collected automatically at 0.3 µm Z-distance using a piezoelectric translator (PIFOC; PI) placed at the base of a 100x PlanApo N.A. 1.4 objective. Fast wavelength changing was achieved by use of a monochromator and a double band-pass filter on the microscope side. The full system was under control of the Metamorph software (Molecular Devices). All experiments were conducted at 37°C, the temperature being controlled by “the Cube” and “the Box” systems (Life Imaging Services, available at www.lis.ch/thebox.html). Automated batch deconvolution of each Z-series was computed using a measured point spread function (PSF) with a custommade software package (J.B. Sibarita, Institut Curie/Section de Recherche). Kymograms along 2D trajectories were used to visualize and quantify vesicle motion along a not labeled cytoskeleton.Citation62 All subsequent image processing and quantifications were done using the ImageJ software (Rasband WS, Image J, US National Institutes of Health, http://rsb.info.nih.gov/ij/, 1997–2009) with the KymoToolBox plugin (F.P. Cordelières, Institut Curie).
Subcellular fractionation and immunoprecipitation.
Cells were harvested 18–20 h after transfection using Trypsin/EDTA, washed with DMEM and homogenized at 4°C in 500 µl of TMD buffer (10 mMTris-HCl pH 7.5, 5 mM MgCl2, 1 mM DTT and a cocktail of protease inhibitors) complemented with 0.25 M sucrose. Nuclei and cellular debris were spun down by centrifugation at 2,500x g for 10 min at 4°C. The resulting supernatant was fractionated by centrifugation at 13,500x g for 30 min at 4°C in order to yield post-Gollgi fractions that retain the Exocyst complex, which was described to sediment by centrifugation at 1,00,000x g in contact-naive cells.Citation63 Immuno-isolation experiments were performed on the supernatant fraction, using Dynal magnetic beads. Dynabeads M280 (Invitrogen, 112-01D) covalently coated with goat anti-mouse IgG, were further coated with 0.4 µg of the primary mouse monoclonal antibody (either anti-Myc, anti-HA or anti-Flag, as indicated) per 107 beads and incubated with subcellular fractions. Finally, bound (B) and unbound (U) components were extracted and subjected to 12.5% SDS-PAGE, electrotransfered onto Hybond ECL nitrocellulose membrane (GE Healthcare Bioscience, RPN78D), and analyzed by immuno-detection using specific primary antibodies as described previously in reference Citation13. For western blots a dilution of I/2,500 was used for anti-Myc and anti-HA antibodies and of 1/1,000 for the anti-FLAG antibody.
Pulldown assays.
MDCK cells were harvested 18–20 h after transfection with indicated plasmids, sedimented, washed with ice-cold PBS containing 5 mM MgCl2 and frozen at −80°C. RhoG activity was determined using a pulldown assay with GST-ELMO1 expressed in Escherichia coli. Bacterial pellets were suspended in ice cold PBS containing a cocktail of protease inhibitors and broken by sonication. After centrifugation at 10,000x g for 20 minutes at 4°C, GST-ELMO1 in the supernatants was purified by incubation with glutathione beads (BD Biosciences, 554780) for 1 hour at 4°C followed by three washes in cold PBS containing protease inhibitors. Pellets of 2 × 107 cells, transfected or not, were lysed by a 20 minutes incubation on ice in lysis buffer, containing 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10 mM MgCl2, 1 mM DTT, 1% Triton x100 and protease inhibitors. The lysate was clarified by centrifugation at 10,000x g for 10 minutes and half of the lysate was used for estimation of total endogenous RhoG and ½ lysate was incubated with GST-ELMO1 beads for 2 h at 4°C, washed three times with lysis buffer and bound proteins were eluted with Laemmli buffer. The eluates and the total cell lysates were separated by SDS-PAGE and probed for RhoG and Flag-Sec3 (1/1,000 dilution for each antibody). A dilution of I/2,500 was used for the anti-Myc antibody.
Yeast two-hybrid and yeast mating assays. The human cDNAs in a brain library fused to Gal4 AD in the two-hybrid vector pAct.2 (a kind gift of Jacques Camonis, Institut Curie, Centre de Recherche, INSERM U528) was used to screen for proteins interacting with the N-terminal peptide of mouse GDI3 (NtGDI3) using conventional two-hybrid techniques (not shown). In brief, yeast reporter strain L40, which contains the reporter genes LacZ and HIS3 downstream of the binding sequences for LexA, was sequentially transformed with the pLexA-NtGDI3 plasmid and with the mouse brain cDNA library in pAct.2 plasmids using the lithium acetate method. The yeast-mating assay was performed using L40 (MATa) and AMR70 (MATα) host strains. Yeasts were transformed according to the lithium acetate protocol and grown at 30°C in appropriate complete dropout medium (DO). The L40 yeast strain was transfected with pFBL23 plasmids containing either mouse or human GDI3 or NtGDI3 cDNAs and grown in DO minus tryptophan. The AMR70 yeast strain was transfected with pAct.2 plasmids containing the cDNAs, found in clones interacting with GDI3Nt and grown in DO minus leucine. L40 transfected cells were mated with AMR70 transfected cells. Diploid cells were selected on DO minus tryptophan and leucine agar and then patched on Whatmann filters to be processed for β-galactosidase activity. Diploids were also tested for histidine prototrophy by selective growth after replication on DO minus tryptophan, leucine and histidine agar. pFBL23 containing the RalB coding sequence and pGAD1318 containing a Ral binding domain (RalBD) sequence were used as positive control vectors, whereas pFBL23 and pGAD1318 without insert were used as negative control vectors. The ability of a LexA-fused protein to interact with a protein fused to the GAL4 activation domain was taken into account only when both LacZ and HIS3 reporter genes were activated in the yeast-mating assay.
Figures and Tables
Figure 1 GDI3 and RhoG interact at the Golgi and on vesicular structures. (A) MDCK cells ectopically expressing GDI3-EYFP alone, showing Golgi and vesicular distribution. (B) MDCK cell expressing EC FP-RhoG alone. Images in (b and c) are shown in black and white mode. RhoG proteins are localized to membrane ruffles (arrows in a and c) and distributed according to a tubulovesicular pattern extending from the perinuclear region throughout the cytoplasm (b). (C and D) Cells co-expressing GDI3-EYFP and EC FP-RhoG. Co-localizations at the Golgi and on vesicular structures are in white in merged image. (E) Bimolecular Fluorescence Complementation (BiFC) assay between RhoG and GDI3. MDCK cells were co-transfected with 20 µg of each YFPNt-RhoG and GDI3-YFPCt plasmids and fixed cells analyzed by confocal microscopy (images a and b). YFP fluorescence is recovered at the Golgi and vesicle-like structures. (F and G) Acceptor photobleaching FRET analysis of RhoG/GDI3 and Rab6/GDI3 interactions. In each experiment, several regions of interest (ROI) at the Golgi region, highlighted by different colors, are shown before and after photobleaching. (F) Cells co-expressing ECFP-RhoG and GDI3-EYFP. FRET efficiencies (%) are 16.18 (green ROI), 16.05 (mauve), 16.74 (yellow), 20.45 (blue), 18.83 (pink) and 17.7 (pale yellow). (G) Cells co-expressing EC FP-Rab6 with GDI3-EYFP, corresponding to the negative control. The calculated FRET efficiencies (%) are 0.12 (green ROI), 0 (mauve), 3.89 (yellow) and 3.44 (blue). All cells were transfected with 20 µg of each indicated plasmid, fixed 20 h post-transfection and analyzed by confocal microscopy. All images represent one single optical section (depth of the field 0.4 µm). Scale bars in µm as indicated.
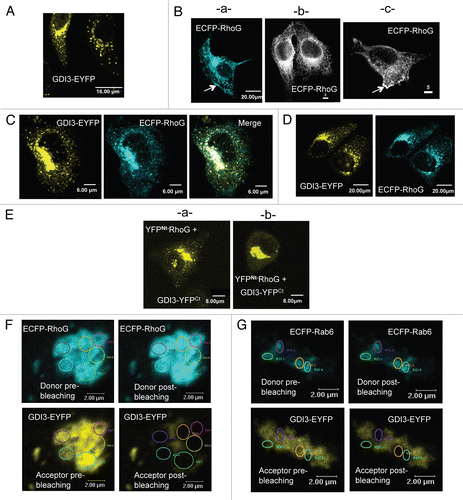
Figure 2 Dynamics of GDI3 and RhoG in live MDCK cells. MDCK cells co-expressing EGFP-RhoG (in green) and GDI3-mCherry (in red) were analyzed by 4D-videomicroscopy. Co-localization appears in yellow in all panels. Scale bar is 5 µm in all parts. (A) Frames taken at 0, 4 and 6 seconds for two different cells. A temporal projection of 10 seconds is shown on the right part. (B) A representative 100-second projection from cell 2 in (A). For raw green and red projections, masks were generated in order to isolate either the RhoG signal alone (a), the GDI3 signal alone (b) or the locations where the two signals co-localize (c). The resulting pseudo-channels display the following features: the EGFP-RhoG traces are mostly located at the cell periphery (arrow in a); GDI3-mCherry traces extend throughout the cell, originating from a peri-centrosomal region (arrow in b); double-labeled structures appear to be larger (arrows in c). The overlay of the 3 pseudo-channels over all time-frames is shown in (d). (C) Representative kymographs generated from the above projection. Left: areas from which the kymographs are generated are shown in white on a 10-second projection. Right: Kymographs generated from a 100-second recording representing the position of RhoG-labeled (1,2), GDI3-labeled (3,4) and double-labeled (5,6) structures, in the horizontal axis, as a function of time, in the vertical axis. Vertical traces correspond to pauses, other traces to movements. Inwards and outwards movements are towards the left and the right, respectively. Speeds are calculated from the slope of the traces. (D) Distribution of speeds of individual structures extracted from the cells shown in (A). The average velocities (µM/min) and standard error of the mean (SEM) are 0.111 (0.039) for Rho-G-labeled structures, 0.108 (0.014) for GDI3-labeled structures and 0.031 (0.005) for double-labeled structures. The average velocity of double-labeled structures is significantly smaller than that of single-labeled RhoG and GDI3 structures (p-values < 0.05 and 0.001, respectively).
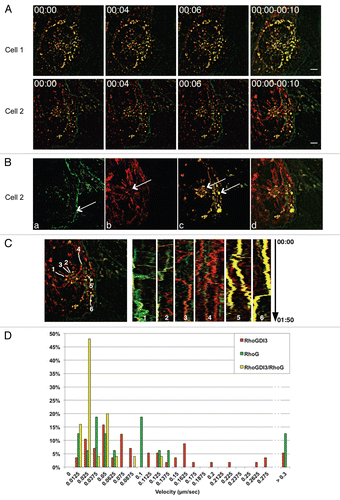
Figure 3 NtGDI3 defines a saturable site at the Golgi. (A) Localization of full-length GDI3-mCherry or NtGDI3-EYFP in MDCK cells. (B) NtGDI3 partially displaces co-expressed GDI3 to the cytoplasm and the cell periphery. MDCK cells are co-transfected with 10 µg of plasmid coding for GDI3-mCherry (red) and 20 µg of NTGDI3-EYFP (green artificial color). The arrow in (Ba) shows patches of GDI3-mCherry at the cell periphery. GDI3-EYFP is also identified at tubulovesicular structures in the cytoplasm (Bb) besides its localization at Golgi and near the plasma membrane. Co-localization at the Golgi is shown in yellow in merged images. (C) NtGDI3 partially displaces co-expressed GDI3 and RhoG to the cytoplasm and the cell periphery in MDCK cells co-expressing GDI3-mCherry, ECFP-RhoG and GDI3Nt-EYFP. Arrows in (Ca and b) show patches of RhoG and GDI3 proteins that co-localize at the cell periphery. The tubulo-vesicular distribution of RhoG and GDI3 is highlighted in Cb. Co-localization of all three proteins appears in white in merged image. (D) MDCK cell co-expressing GDI3-mCherry (red) and EC FP-RhoG (green artificial color) in the absence of NtGDI3-EYFP. Both proteins colocalize at Golgi and vesicles (yellow). Cells were fixed 20 h post-transfection and analyzed by confocal microscopy. Scale bars are in µm as indicated.
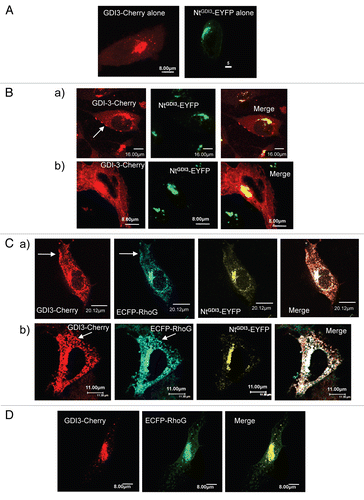
Figure 4 GDI3 interacts with Sec3 and recruits it to Golgi and vesicles. (A) Detection of cytoplasmic complexes between Flag-Sec3 and GDI3-HA by co-immunoprecipitation (IP). HeLa cells were co-transfected with 20 µg of each plasmid. Post-Golgi supernatant fractions (see Materials and Methods) were immunoprecipitated with magnetic beads coated with either anti-HA (HA ) or anti-Flag (Flag) monoclonal antibody. Bound (B) and unbound (U) materials were collected and analyzed by western blotting. (B) Identification of GDI3/Sec3 complexes at the Golgi apparatus by the BiFC assay. MDCK cells were co-transfected with equal amounts (20 µg) of YFPNt-Sec3 and GDI3-YFPCt coding plasmids. Yellow fluorescence is recovered at Golgi structures. (C) ECFP-Sec3 is distributed diffusely throughout the cytoplasm in MDCK cells (images a and b). Cells were transfected with plasmids coding for ECFP-Sec3 (20 µg). (D) MDCK cells co-expressing ECFP-Sec3 and GDI3-EYFP showing that GDI3-EYFP displaces ECFP-Sec3 from the cytoplasm to perinuclear and vesicular structures. (E) MCDK cells co-expressing ECFP-Sec3 and NtGDI3-EYFP (A and B). The N-terminal extension of GDI3 is sufficient to target Sec3 to Golgi and vesicular structures. Co-localization at the Golgi and on vesicular structures appears in white in merged images. Cells in (B–F) were analyzed by confocal microscopy. All images represent one single optical section (depth of the field 0.4 µm). Scale bars are in µm as indicated.
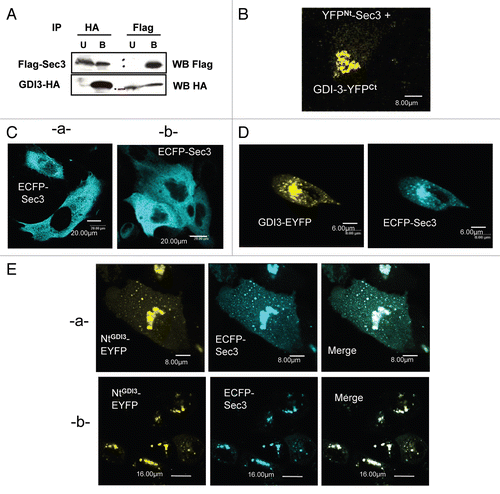
Figure 5 GDI3 recruits the Sec8 Exocyst subunit to Golgi and vesicles in MDCK cells but not the Sec6 subunit. (A) Endogenous Sec8 has a diffuse cytoplasmic localization in non-confluent monolayers of MDCK cells. (B) Endogenous Sec8 retains this diffuse cytoplasmic localization in cells expressing ECFP-Sec3. (C) GDI3-EYFP (highlighted in green artificial color) targets endogenous Sec8 to Golgi and vesicles. Co-localized proteins appear in yellow in merge image. (D) GDI3-EYFP co-expressed with ECFP-Sec3 targets both ECFP-Sec3 and the endogenous Sec8 protein to Golgi and vesicular structures. (E) Endogenous Sec6 localizes at the cell periphery and at some punctuate structures in the cytoplasm. (F) Expression of GDI3-EYFP has no effect on the plasma membrane localization of Sec6 in MDCK cells. (G) Co-expression of GDI3-EYFP with ECFP-Sec3 is also ineffective at displacing Sec6 from the plasma membrane. GDI3-EYFP is shown in green artificial color. In all panels mouse anti-Sec8 monoclonal antibody and mouse anti-Sec6 monoclonal antibody were used at a dilution of 1/200 and the secondary anti-mouse antibody Alexa568 at 1/800. Scale bars in µm as indicated.
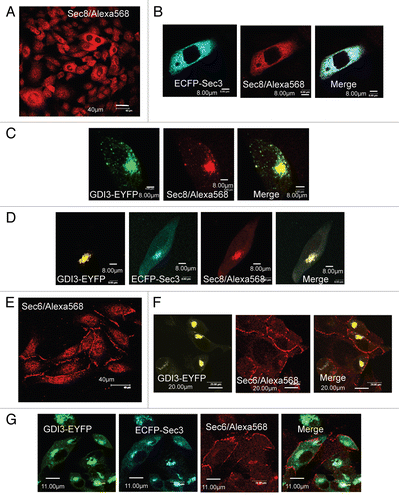
Figure 6 Sec3 induces TNT-like membrane protrusions and increases the level of RhoG-GTP in MDCK cells. (A) EGFP-RhoG localizes to perinuclear structures, to vesicles and to the plasma membrane where it induces membrane ruffles (images a and b). (B) Co-expression of ECFP-Sec3 with EGFP-RhoG induces TNT-like structures in MDCK cells. EGFP-RhoG and ECFP-Sec3 co-localize at perinuclear structures, at tubulovesicular cytoplasmic structures and in TNT-like protrusions connecting neighboring cells (arrows in a–c). Scale bars in µm as indicated. (C) Transient expression of Flag-Sec3 in MDCK cells increases the level of GTP-bound endogenous RhoG. Endogenous RhoG activity in lysed MDCK cells was measured in supernatants after centrifugation at 10,000x g by pull-down assays using GST-ELMO and normalized to total endogenous RhoG (relative levels of active RhoG). Western blots showing the active RhoG levels: (lane 1) MDCK cells transiently expressing GDI3 show very low levels of active endogenous RhoG-GTP; (lane 2) MDCK cells transiently expressing Flag-Sec3 show a 15-fold increase of endogenous RhoG-GTP compared to the relative RhoG-GTP levels in non-transfected MDCK cells; (lane 3) non-transfected MDCK cells show low level of RhoG-GTP. Flag-Sec3 and endogenous RhoG are highlighted in western blots, using a mixture of anti-Flag and anti-RhoG antibodies.
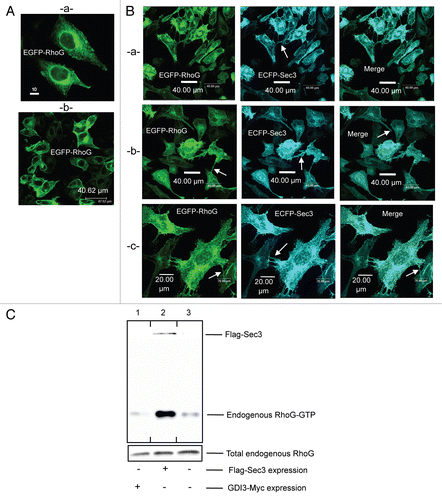
Figure 7 Downregulation of RhoG and Sec3 functions by GDI3. (A) GDI3 relocates both RhoG and Sec3 to the Golgi apparatus and suppresses membrane protrusions and ruffles. Triple transfection of GDI3-mCherry, EGFP-RhoG and ECFP-Sec3 proteins in MDCK cells are done with 15 µg of each plasmid. Co-localization is seen in white in merged image. Scale bars in µm as indicated. (B) ECFP-RhoG displaces the endogenous Sec8 Exocyst subunit from the cytoplasm to the cell periphery (see arrows) and to the Golgi. (C) GDI3-EYFP co-expressed with ECFP-RhoG targets these proteins and the endogenous Sec8 protein to Golgi and vesicular structures. (D) Co-expression of GDI3-EYFP with ECFP-RhoG has no effect on the plasma membrane localization of Sec6 in MDCK cells. Mouse anti-Sec8 monoclonal antibody and mouse anti-Sec6 monoclonal antibody in (C–E) were used at a dilution of 1/200 and the secondary anti-mouse antibody Alexa568 at 1/800. Scale bars in µm as indicated.
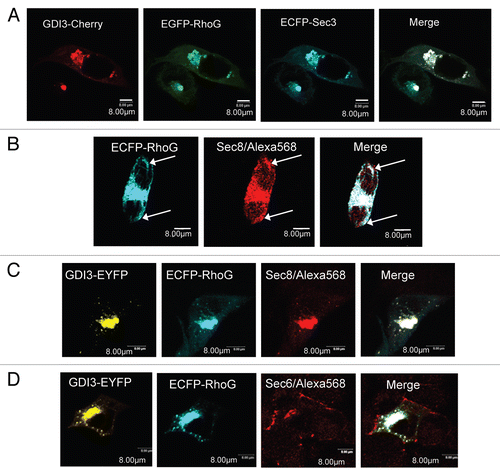
Table 1 Yeast two-hybrid mating assay
Additional material
Download Zip (59.8 MB)Acknowledgements
This work was supported by the CNRS, by grants from the French Ministry of Research (Action Concertée Incitative-Biologie cellulaire, Moléculaire et Structurale), and the Association pour la Recherche contre le Cancer (Grant number 3234). We thank Susanne Bolte, Marie-Noelle Soler, Monica Debreczeny, Spencer Brown and all the staff at the Cell Imaging Facility (IMAGIF), Centre de Recherche de Gif, CNRS, Gif-sur-Yvette, France for their warm welcome and invaluable help for all confocal microscopy studies. We are also grateful to all the staff of “Petites ProtéinesG: Structures, Interactions et Régulations cellulaires a at LEBS, CNRS, Gif-sur-Yvette, France.
References
- Ridley AJ. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol 2006; 16:522 - 529
- Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol 2008; 9:690 - 701
- Sahai E, Marshall CJ. RHO-GTPases and cancer. Nat Rev Cancer 2002; 2:133 - 142
- Vega FM, Ridley AJ. Rho GTPases in cancer cell biology. FEBS Lett 2008; 582:2093 - 2101
- Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy?. Nat Rev Cancer 2010; 10:842 - 857
- Olofsson B. Rho guanine dissociation inhibitors: Pivotal molecules in cellular signalling. Cell Signal 1999; 11:545 - 554
- DerMardirossian C, Bokoch GM. GDIs: central regulatory molecules in Rho GTPase activation. Trends Cell Biol 2005; 15:356 - 363
- Dransart E, Olofsson B, Cherfils J. RhoGDIs Revisited: Novel Roles in Rho Regulation. Traffic 2005; 6:957 - 966
- Theodorescu D, Sapinoso LM, Conaway MR, Oxford G, Hampton GM, Frierson HF Jr. Reduced expression of metastasis suppressor RhoGDI2 is associated with decreased survival for patients with bladder cancer. Clin Cancer Res 2004; 10:3800 - 3806
- Zhang B. Rho GDP dissociation inhibitors as potential targets for anticancer treatment. Drug Resist Updat 2006; 9:134 - 141
- Wu Y, McRoberts K, Berr SS, Frierson HF Jr, Conaway M, Theodorescu D. Neuromedin U is regulated by the metastasis suppressor RhoGDI2 and is a novel promoter of tumor formation, lung metastasis and cancer cachexia. Oncogene 2006; 26:765 - 773
- Zalcman G, Closson V, Camonis J, Honore N, Rousseau-Merck MF, Tavitian A, et al. RhoGDI-3 is a new GDP dissociation inhibitor (GDI). Identification of a non-cytosolic GDI protein interacting with the small GTP-binding proteins RhoB and RhoG. J Biol Chem 1996; 271:30366 - 30374
- Brunet N, Morin A, Olofsson B. RhoGDI-3 regulates RhoG and targets this protein to the Golgi complex through its unique N-terminal domain. Traffic 2002; 3:342 - 358
- Dransart E, Morin A, Cherfils J, Olofsson B. Uncoupling of inhibitory and shuttling functions of rho GDP dissociation inhibitors. J Biol Chem 2005; 280:4674 - 4683
- Jiang WG, Watkins G, Lane J, Cunnick GH, Douglas-Jones A, Mokbel K, et al. Prognostic value of rho GTPases and rho guanine nucleotide dissociation inhibitors in human breast cancers. Clin Cancer Res 2003; 9:6432 - 6440
- Blangy A, Vignal E, Schmidt S, Debant A, Gauthier-Rouviere C, Fort P. TrioGEF1 controls Rac- and Cdc42-dependent cell structures through the direct activation of rhoG. J Cell Sci 2000; 113:729 - 739
- Vignal E, Blangy A, Martin M, Gauthier-Rouviere C, Fort P. Kinectin is a key effector of RhoG microtubule-dependent cellular activity. Mol Cell Biol 2001; 21:8022 - 8034
- Prieto-Sanchez RM, Berenjeno IM, Bustelo XR. Involvement of the Rho/Rac family member RhoG in caveolar endocytosis. Oncogene 2006; 25:2961 - 2973
- Katoh H, Negishi M. RhoG activates Rac1 by direct interaction with the Dock180-binding protein Elmo. Nature 2003; 424:461 - 464
- Katoh H, Yasui H, Yamaguchi Y, Aoki J, Fujita H, Mori K, et al. Small GTPase RhoG is a key regulator for neurite outgrowth in PC12 cells. Mol Cell Biol 2000; 20:7378 - 7387
- Brugnera E, Haney L, Grimsley C, Lu M, Walk SF, Tosello-Trampont AC, et al. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat Cell Biol 2002; 4:574 - 582
- Hiramoto K, Negishi M, Katoh H. Dock4 is regulated by RhoG and promotes Rac-dependent cell migration. Exper Cell Res 2006; 312:4205 - 4216
- Katoh H, Hiramoto K, Negishi M. Activation of Rac1 by RhoG regulates cell migration. J Cell Sci 2006; 119:56 - 65
- Tosello-Trampont AC, Kinchen JM, Brugnera E, Haney LB, Hengartner MO, Ravichandran KS. Identification of two signaling submodules within the CrkII/ELMO/Dock180 pathway regulating engulfment of apoptotic cells. Cell Death Differ 2007; 14:963 - 972
- D'Angelo R, Aresta S, Blangy A, Del Maestro L, Louvard D, Arpin M. Interaction of ezrin with the novel guanine nucleotide exchange factor PLEKHG6 promotes RhoG-dependent apical cytoskeleton rearrangements in epithelial cells. Mol Biol Cell 2007; 18:4780 - 4793
- Bouquier N, Vignal E, Charrasse S, Weill M, Schmidt S, LÈonetti JP, et al. A cell active chemical GEF inhibitor selectively targets the Trio/RhoG/Rac1 signaling pathway. Chem Biol 2009; 16:657 - 666
- Ellerbroek SM, Wennerberg K, Arthur WT, Dunty JM, Bowman DR, DeMali KA, et al. SGEF, a RhoG guanine nucleotide exchange factor that stimulates macropinocytosis. Mol Biol Cell 2004; 15:3309 - 3319
- deBakker CD, Haney LB, Kinchen JM, Grimsley C, Lu M, Klingele D, et al. Phagocytosis of apoptotic cells is regulated by a UNC-73/TRIO-MIG-2/RhoG signaling module and armadillo repeats of CED-12/ELMO. Curr Biol 2004; 14:2208 - 2216
- Nakaya M, Tanaka M, Okabe Y, Hanayama R, Nagata S. Opposite effects of rho family GTPases on engulfment of apoptotic cells by macrophages. J Biol Chem 2006; 281:8836 - 8842
- van Buul JD, Allingham MJ, Samson T, Meller J, Boulter E, Garcia-Mata R, et al. RhoG regulates endothelial apical cup assembly downstream from ICAM1 engagement and is involved in leukocyte trans-endothelial migration. J Cell Biol 2007; 178:1279 - 1293
- Samson T, Welch C, Monaghan-Benson E, Hahn KM, Burridge K. Endogenous RhoG is rapidly activated after epidermal growth factor stimulation through multiple guanine-nucleotide exchange factors. Mol Biol Cell 2010; 21:1629 - 1642
- Tu S, Wu WJ, Wang J, Cerione RA. Epidermal growth factor-dependent regulation of Cdc42 is mediated by the Src tyrosine kinase. J Biol Chem 2003; 278:49293 - 49300
- DerMardirossian C, Rocklin G, Seo JY, Bokoch GM. Phosphorylation of RhoGDI by Src regulates Rho GTPase binding and cytosol-membrane cycling. Mol Biol Cell 2006; 17:4760 - 4768
- Elfenbein A, Rhodes JM, Meller J, Schwartz MA, Matsuda M, Simons M. Suppression of RhoG activity is mediated by a syndecan 4-synectin-RhoGDI1 complex and is reversed by PKC{alpha} in a Rac1 activation pathway. J Cell Biol 2009; 186:75 - 83
- Groysman M, Hornstein I, Alcover A, Katzav S. Vav1 and Ly-GDI two regulators of Rho GTPases, function cooperatively as signal transducers in T cell antigen receptor-induced pathways. J Biol Chem 2002; 277:50121 - 50130
- Robbe K, Otto-Bruc A, Chardin P, Antonny B. Dissociation of GDP dissociation inhibitor and membrane translocation are required for efficient activation of Rac by the Dbl homology-pleckstrin homology region of Tiam. J Biol Chem 2003; 278:4756 - 4762
- Ugolev Y, Berdichevsky Y, Weinbaum C, Pick E. Dissociation of Rac1(GDP): RhoGDI complexes by the cooperative action of anionic liposomes containing phosphatidylinositol-3,4,5-trisphosphate, Rac guanine nucleotide exchange factor and GTP. J Biol Chem 2008; 283:22257 - 22571
- Johnson JL, Erickson JW, Cerione RA. New insights into how the Rho guanine nucleotide dissociation inhibitor regulates the interaction of Cdc42 with membranes. J Biol Chem 2009; 284:23860 - 23871
- Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 2002; 9:789 - 798
- He L, Bradrick TD, Karpova TS, Wu X, Fox MH, Fischer R, et al. Flow cytometric measurement of fluorescence (Förster) resonance energy transfer from cyan fluorescent protein to yellow fluorescent protein using single-laser excitation at 458 nm. Cytometry A 2003; 53:39 - 54
- Matern HT, Yeaman C, Nelson WJ, Scheller RH. Inaugural Article: The Sec6/8 complex in mammalian cells: Characterization of mammalian Sec3, subunit interactions and expression of subunits in polarized cells. Proc Natl Acad Sci 2001; 98:9648 - 9653
- Hsu SC, TerBush D, Abraham M, Guo W, Kwang WJ. The exocyst complex in polarized exocytosis. Int Rev Cytol 2004; 233:243 - 265
- Wang S, Hsu SC. The molecular mechanisms of the mammalian exocyst complex in exocytosis. Biochem Soc Trans 2006; 34:687 - 690
- Yeaman C, Grindstaff KK, Wright JR, Nelson WJ. Sec6/8 complexes on trans-Golgi network and plasma membrane regulate late stages of exocytosis in mammalian cells. J Cell Biol 2001; 155:593 - 604
- Vik-Mo EO, Oltedal L, Hoivik EA, Kleivdal H, Eidet J, Davanger S. Sec6 is localized to the plasma membrane of mature synaptic terminals and is transported with secretogranin ii-containing vesicles. Neuroscience 2003; 119:73 - 85
- Yeaman C, Grindstaff KK, Nelson WJ. Mechanism of recruiting Sec6/8 (exocyst) complex to the apical junctional complex during polarization of epithelial cells. J Cell Sci 2004; 117:559 - 570
- Oztan A, Silvis M, Weisz OA, Bradbury NA, Hsu SC, Goldenring JR, et al. Exocyst Requirement for Endocytic Traffic Directed Toward the Apical and Basolateral Poles of Polarized MDCK Cells. Mol Biol Cell 2007; 18:3978 - 3992
- Sakurai-Yageta M, Recchi C, Le Dez G, Sibarita JB, Daviet L, Camonis J, et al. The interaction of IQGAP1 with the exocyst complex is required for tumor cell invasion downstream of Cdc42 and RhoA. J Cell Biol 2008; 181:985 - 998
- Rustom A, Saffrich R, Markovic I, Walther P, Gerdes HH. Nanotubular Highways for Intercellular Organelle Transport. Science 2004; 303:1007 - 1010
- Gerdes HH, Bukoreshtliev NV, Barroso JoFV. Tunneling nanotubes: A new route for the exchange of components between animal cells. FEBS Lett 2007; 581:2194 - 2201
- Hase K, Kimura S, Takatsu H, Ohmae M, Kawano S, Kitamura H, et al. M-Sec promotes membrane nanotube formation by interacting with Ral and the exocyst complex. Nat Cell Biol 2009; 11:1427 - 1432
- Munson M, Novick P. The exocyst defrocked, a framework of rods revealed. Nat Struct Mol Biol 2006; 13:577 - 581
- He B, Guo W. The exocyst complex in polarized exocytosis. Curr Opin Cell Biol 2009; 21:537 - 542
- Guo W, Tamanoi F, Novick P. Spatial regulation of the exocyst complex by Rho1 GTPase. Nat Cell Biol 2001; 3:353 - 360
- Yamashita M, Kurokawa K, Sato Y, Yamagata A, Mimura H, Yoshikawa A, et al. Structural basis for the Rho- and phosphoinositide-dependent localization of the exocyst subunit Sec3. Nat Struct Mol Biol 2010; 17:180 - 186
- Rosse C, Hatzoglou A, Parrini MC, White MA, Chavrier P, Camonis J. RalB Mobilizes the exocyst to drive cell migration. Mol Cell Biol 2006; 26:727 - 734
- Zuo X, Zhang J, Zhang Y, Hsu SC, Zhou D, Guo W. Exo70 interacts with the Arp2/3 complex and regulates cell migration. Nat Cell Biol 2006; 8:1383 - 1388
- Letinic K, Sebastian R, Toomre D, Rakic P. Exocyst is involved in polarized cell migration and cerebral cortical development. Proc Natl Acad Sci USA 2009; 106:11342 - 11347
- Morin A, Picart R, Tixier-Vidal A. Effects of the N-terminal cysteine mutation on prolactin expression and secretion in transfected pituitary cells. Mol Cell Endocrinol 1996; 117:59 - 73
- Wilson CGM, Magliery TJ, Regan L. Detecting protein-protein interactions with GFP-fragment reassembly. Nat Meth 2004; 1:255 - 262
- Kerppola TK. Bimolecular fluorescence complementation: visualization of molecular interactions in living cells. Methods Cell Biol 2008; 85:431 - 470
- Racine V, Sachse M, Salamero J, Fraisier V, Trubuil A, Sibarita JB. Visualization and quantification of vesicle trafficking on a three-dimensional cytoskeleton network in living cells. J Microsc 2007; 225:214 - 228
- Yeaman C. Ultracentrifugation-based approaches to study regulation of Sec6/8 (exocyst) complex function during development of epithelial cell polarity. Methods 2003; 30:198 - 206