Abstract
Rictor’s role in cell migration has been first indicated in the original chemotaxis studies in Dictyostelium and more recent studies reported that rictor is required for migration of cancer cells. How rictor promotes cell migration remains poorly characterized. Based on our proteomics study we have identified a novel functional role of rictor in regulation of cell migration. Here, we discuss our recent finding that rictor by suppressing RhoGDI2 maintains activity of the Rac1/cdc42 GTPases and promotes cell migration. Our finding outlines a critical role of rictor in the regulation of RhoGDI2 activity. This study opens new avenues in the investigation of cancer metastasis by analyzing the rictor dependent post-translational modification of RhoGDI2.
Keywords: :
Directed cell migration is a fundamental cellular response required for embryonic development, inflammatory response and wound repair.Citation1 The Rho GTPases provide major role in coordinating the cellular responses required for cell migration. Rho proteins act as molecular switches cycling between an active GTP-bound and inactive GDP-bound conformations. This activity is tightly controlled by the guanine nucleotide exchange factors and GTPase activating proteins that determine a ratio of active or inactive Rho GTPases in cells.Citation2 Rho specific guanine nucleotide dissociation inhibitors (RhoGDIs) represent additional class of regulatory proteins that are critical to control activity of Rho GTPases. By forming heterodimers with the inactive GDP-bound Rho GTPases, the RhoGDI proteins control the Rho GTPase partitioning between the cytosol and membrane compartments.Citation3
In mammals, the RhoGDI family is represented by three members, each member displays a specific pattern of tissue distribution and activity toward Rho GTPases.Citation2 RhoGDI1, as the most abundant and ubiquitously expressed member of the RhoGDI family, has been actively studied and characterized.Citation2 Similar to RhoGDI1, the second member RhoGDI2 is also a conserved member of this family and plays an important role in cell migration.Citation4-Citation10 The third member of this family RhoGDI3 is a least conserved member of RhoGDIs and known to be expressed preferentially in brain, pancreas, lung, kidney and testis.Citation11
The initial characterization of RhoGDI2 identified its tissue specific expression in the blood forming cells, but the later study determined its expression in a wide range of cell types.Citation5 RhoGDI2 along with RhoGDI1 share common structural features and interact with the same and distinct Rho GTPases to regulate their membrane association.Citation12 Despite a high degree of structure similarity, some studies suggest that both RhoGDIs have specific binding affinities toward active Rho GTPases.Citation13-Citation15 RhoGDI1 preferentially binds and inhibits RhoA,(13,15) whereas RhoGDI2 has been shown to bind and inhibit Rac GTPase.Citation14 Remarkably, RhoGDI2 has been identified as a marker lost in bladder cancer metastasis and its loss correlates with a decreased patient survival.Citation5 Similarly, others have reported that abundance of RhoGDI2 is either decreased or lost in several invasive cancers.Citation4,Citation7,Citation9 Given the increasing evidence regarding a loss of RhoGDI2 in cancer metastasis, it is important to explore the molecular mechanism of regulation of RhoGDI2 and its implication in cancer cell migration and metastasis. Our recent study presented in Oncogene reveals that rictor (rapamycin-insensitive companion of mTOR) controls the functional activity of RhoGDI2.Citation16
Initially, the Devreotes group has identified the rictor’s ortholog in Dictyostelium as a regulator of chemotaxis.Citation17 Later, the biochemical studies of the mammalian Target of Rapamycin (mTOR), the essential protein kinase known as a central component of the highly conserved nutrient-sensing pathway, defined rictor as the mTOR interacting protein.Citation18,Citation19 Binding of rictor and Sin1 to mTOR forms mTORC2 (the mTOR Complex 2) distinct from the complex assembled by mTOR and raptor (mTORC1). mTORC1 is a nutrient-sensitive complex that regulates protein synthesis by phosphorylating the regulators of protein synthesis S6K1 and 4EBP1.Citation20 The second mTOR complex, mTORC2 functions as the regulatory kinase of the distinct members of AGC kinase family including Akt, SGK and PKCα. In yeast and mammalian cells, mTORC2 has been shown to regulate cell morphology, at least in part, by controlling protein kinase Cα18 and activity of the Rho GTPases.Citation19,Citation21
The functional studies of rictor have been mostly focused on its role as the essential component of mTORC2. The recent studies are also beginning to shed the light on a role of rictor in cell migration independent of mTORC2. It has been shown that rictor alone without its binding partner Sin1 interacts with the regulators of cell morphology and migration known as the integrin-linked kinase and actin-based molecular motor myosin-1c.Citation22,Citation23 In our recent work, we found that a loss of rictor leads to induction of RhoGDI2 and interferes with cell migration by inhibition of Rac1 and cdc42 GTPase activity. Importantly, the rictor binding protein Sin1, an integral component of mTORC2, has not been linked to the regulation of RhoGDI2 indicating a novel mTORC2 independent function of rictor in cell migration.Citation16
To address a functional role of rictor in regulation of cell migration, we performed an unbiased proteomic analysis of the rictor null mouse embryonic fibroblasts (MEFs). We identified several differentially expressed proteins correspond to actin filament assembly and stress response pathways.Citation16 Cofilin and destrin were shown to play an important role in the assembly of actin filament by forming new polymerization-competent filament barbed ends through their severing activityCitation24 were low expressed proteins in the rictor null cells. These differences might reflect a lower cytoskeleton dynamics of the elongated rictor null cells.Citation16 On the contrary, we also observed a high abundance of RhoGDI2 in the rictor null cells,Citation16 which is known to function as an important negative regulator of Rho GTPases.Citation2
RhoGDI2 is highly conserved protein containing two structurally distinct regions. The N-terminal flexible region (residues 1–74) defines its “regulatory arm” and is responsible for the GTPase inhibitory function through interactions with the Rac GTPase, whereas the C-terminal region (residues 75–204) accommodates the isoprenyl moiety of the Rac GTPase in its hydrophobic pocket, and is required to extract Rac GTPases from the membrane (). Therefore, both regions of RhoGDI2 contribute significantly in the Rac binding and the GTPase activity of the Rac.Citation14 Our finding is consistent with the previous studies indicating that RhoGDI2 interacts with Rac1 and Cdc42 but with a higher affinity toward Rac1 carrying a substantial inhibitory effect on GTPase activity of Rac1.Citation16 Thus, the association of RhoGDI2-Rac1 GTPase complex is likely plays an important role in the regulation of Rac1 GTPase activity. A key question remains to be answered is how rictor regulates RhoGDI2. This regulation is not related to the gene expression mechanism because overexpression of the wild type RhoGDI2 does not mimic the effect of a loss of rictor in the wild type MEFs, although a suppression of RhoGDI2 in the rictor null MEFs is effective in rescue of cell migration.Citation16 It is likely to be mediated by a rictor dependent post-translational modification of RhoGDI2 at the extreme N-terminal regulatory sequence.
Figure 1. (A) Structure of RhoGDI1 and RhoGDI2. Primary sequence similiarity of RhoGDI1 and RhoGDI2 as indicated by the percent identity. The approximate locations of important residues described in the text are indicated the terminal amino acid residue. First 25 amino acid residues are defined as extreme N-terminal region. (B) Sequence alignments of human RhoGDI1 and RhoGDI2 were performed using COBALT FORMAT. *Indicated the possible post-translational modication residues.
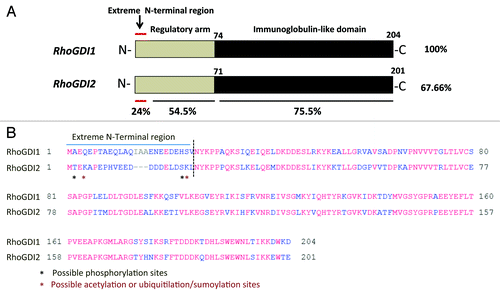
The importance of the functional role of the N-terminal region of RhoGDI2 has been previously described with truncated N-terminally proteins.Citation25 Deletion of the first 41 amino acid residues from RhoGDI2 resulted in complete loss of its ability to bind Rac1 and GTPase activity.Citation25 Although the folded regions of the RhoGDI1 and RhoGDI2 show 74% sequence identity, whereas the N-terminal sequence, with the first 25 amino acid residues show only 24% of identity and the fragment representing residues between amino acids 26–74 carries identity of 54.5% (). Therefore, we assume that rictor alone forms a distinct complex with the unknown kinase and regulate the phosphorylation in the extreme N-terminal amino acid residues that varies between RhoGDI1 and RhoGDI2 (). Phosphorylation of RhoGDIs has emerged as one of the key post-translational modifications that may play a major role in the dissociation of the cytosolic RhoGDIs-GTPase complexes.Citation26-Citation29 Several phosphorylation sites have been identified on RhoGDI1 such as Ser101 and Ser174 phosphorylation by p-21-activated kinaseCitation30 and Ser34 by protein kinase Cα (PKCα)Citation28 promoting the release of RhoA and Rac1 respectively. Recently, similar phosphorylation site (Ser31) by PKCα was identified on RhoGDI2.Citation29 Griner and his colleagues reported that PKCα-mediated phosphorylation of Ser31 destabilizes the N-terminus of the RhoGDI2 that interacts mainly with the switch regions of Rac1 GTPases most likely accounting for the decrease in RhoGDI2 binding to Rac1 and promotes its GTPase activity.Citation29 Therefore it is also possible that the functional activity of RhoGDI2 controlled by rictor depends on other type of the post-translational modifications (the extreme N-terminal regulatory region) such as acetylation or ubiquitilation/sumoylation () because the RhoGDI1 sumoylation at Lys-138 has been shown to be crucial for cell migration.Citation31
RhoGDI2 has been actively studied in cancer metastasis and the expression levels of RhoGDI2 are severely altered in a wide range of cancers.Citation4-Citation10 It has been reported that the abundance of RhoGDI2 is either decreased or lost in several cancers.Citation4,Citation5,Citation7,Citation9,Citation10 In contrary, the studies in pancreatic cancer reported a high abundance of RhoGDI2 and it correlates with increased invasiveness.Citation8 In breast cancer, the pattern of expression of RhoGDI2 is biphasic. There is evidence that the expression of RhoGDI2 is increased at the early stages of cancer progression but decreased sharply during metastasis.Citation6 We believe that these studies by focusing on detection of the RhoGDI2 abundance as a marker do not reflect the functional activity of RhoGDI2. Most likely, the functional role of RhoGDI2 as a potent inhibitor of cell migration is dependent on its activity regulated by phosphorylation or other posttranslational modifications within extreme N-terminal domain because it is distinct from RhoGDI1. Overall, our study points out on the potent rictor dependent regulation of the RhoGDI2 functional activity and understanding of this mechanism carries a promise to translate into the novel approach to control cell migration and treat cancer metastasis.
Acknowledgments
This study was supported by the MD Anderson Metastasis Research Center and the NIH grants CA133522 (D.D.S.).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Lauffenburger DA, Horwitz AF. Cell migration: a physically integrated molecular process. Cell 1996; 84:359 - 69; http://dx.doi.org/10.1016/S0092-8674(00)81280-5; PMID: 8608589
- Dovas A, Couchman JR. RhoGDI: multiple functions in the regulation of Rho family GTPase activities. Biochem J 2005; 390:1 - 9; http://dx.doi.org/10.1042/BJ20050104; PMID: 16083425
- Nomanbhoy TK, Cerione R. Characterization of the interaction between RhoGDI and Cdc42Hs using fluorescence spectroscopy. J Biol Chem 1996; 271:10004 - 9; http://dx.doi.org/10.1074/jbc.271.17.10004; PMID: 8626553
- Titus B, Frierson HF Jr., Conaway M, Ching K, Guise T, Chirgwin J, et al. Endothelin axis is a target of the lung metastasis suppressor gene RhoGDI2. Cancer Res 2005; 65:7320 - 7; http://dx.doi.org/10.1158/0008-5472.CAN-05-1403; PMID: 16103083
- Theodorescu D, Sapinoso LM, Conaway MR, Oxford G, Hampton GM, Frierson HF Jr.. Reduced expression of metastasis suppressor RhoGDI2 is associated with decreased survival for patients with bladder cancer. Clin Cancer Res 2004; 10:3800 - 6; http://dx.doi.org/10.1158/1078-0432.CCR-03-0653; PMID: 15173088
- Hu LD, Zou HF, Zhan SX, Cao KM. Biphasic expression of RhoGDI2 in the progression of breast cancer and its negative relation with lymph node metastasis. Oncol Rep 2007; 17:1383 - 9; PMID: 17487395
- Gildea JJ, Seraj MJ, Oxford G, Harding MA, Hampton GM, Moskaluk CA, et al. RhoGDI2 is an invasion and metastasis suppressor gene in human cancer. Cancer Res 2002; 62:6418 - 23; PMID: 12438227
- Abiatari I, DeOliveira T, Kerkadze V, Schwager C, Esposito I, Giese NA, et al. Consensus transcriptome signature of perineural invasion in pancreatic carcinoma. Mol Cancer Ther 2009; 8:1494 - 504; http://dx.doi.org/10.1158/1535-7163.MCT-08-0755; PMID: 19509238
- Jiang WG, Watkins G, Lane J, Cunnick GH, Douglas-Jones A, Mokbel K, et al. Prognostic value of rho GTPases and rho guanine nucleotide dissociation inhibitors in human breast cancers. Clin Cancer Res 2003; 9:6432 - 40; PMID: 14695145
- Ma L, Xu G, Sotnikova A, Szczepanowski M, Giefing M, Krause K, et al. Loss of expression of LyGDI (ARHGDIB), a rho GDP-dissociation inhibitor, in Hodgkin lymphoma. Br J Haematol 2007; 139:217 - 23; http://dx.doi.org/10.1111/j.1365-2141.2007.06782.x; PMID: 17897297
- Dransart E, Morin A, Cherfils J, Olofsson B. RhoGDI-3, a promising system to investigate the regulatory function of rhoGDIs: uncoupling of inhibitory and shuttling functions of rhoGDIs. Biochem Soc Trans 2005; 33:623 - 6; http://dx.doi.org/10.1042/BST0330623; PMID: 16042558
- Garcia-Mata R, Boulter E, Burridge K. The ‘invisible hand’: regulation of RHO GTPases by RHOGDIs. Nat Rev Mol Cell Biol 2011; 12:493 - 504; http://dx.doi.org/10.1038/nrm3153; PMID: 21779026
- Longenecker K, Read P, Derewenda U, Dauter Z, Liu X, Garrard S, et al. How RhoGDI binds Rho. Acta Crystallogr D Biol Crystallogr 1999; 55:1503 - 15; http://dx.doi.org/10.1107/S090744499900801X; PMID: 10489445
- Scheffzek K, Stephan I, Jensen ON, Illenberger D, Gierschik P. The Rac-RhoGDI complex and the structural basis for the regulation of Rho proteins by RhoGDI. Nat Struct Biol 2000; 7:122 - 6; http://dx.doi.org/10.1038/72392; PMID: 10655614
- Tnimov Z, Guo Z, Gambin Y, Nguyen UT, Wu YW, Abankwa D, et al. Quantitative analysis of prenylated RhoA interaction with its chaperone, RhoGDI. J Biol Chem 2012; 287:26549 - 62; http://dx.doi.org/10.1074/jbc.M112.371294; PMID: 22628549
- Agarwal NK, Chen CH, Cho H, Boulbès DR, Spooner E, Sarbassov DD. Rictor regulates cell migration by suppressing RhoGDI2. Oncogene 2012; In press http://dx.doi.org/10.1038/onc.2012.287; PMID: 22777355
- Chen MY, Long Y, Devreotes PN. A novel cytosolic regulator, Pianissimo, is required for chemoattractant receptor and G protein-mediated activation of the 12 transmembrane domain adenylyl cyclase in Dictyostelium. Genes Dev 1997; 11:3218 - 31; http://dx.doi.org/10.1101/gad.11.23.3218; PMID: 9389653
- Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol 2004; 14:1296 - 302; http://dx.doi.org/10.1016/j.cub.2004.06.054; PMID: 15268862
- Jacinto E, Loewith R, Schmidt A, Lin S, Rüegg MA, Hall A, et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol 2004; 6:1122 - 8; http://dx.doi.org/10.1038/ncb1183; PMID: 15467718
- Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol 2005; 17:596 - 603; http://dx.doi.org/10.1016/j.ceb.2005.09.009; PMID: 16226444
- Schmidt A, Bickle M, Beck T, Hall MN. The yeast phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell 1997; 88:531 - 42; http://dx.doi.org/10.1016/S0092-8674(00)81893-0; PMID: 9038344
- Hagan GN, Lin Y, Magnuson MA, Avruch J, Czech MPA. A Rictor-Myo1c complex participates in dynamic cortical actin events in 3T3-L1 adipocytes. Mol Cell Biol 2008; 28:4215 - 26; http://dx.doi.org/10.1128/MCB.00867-07; PMID: 18426911
- McDonald PC, Oloumi A, Mills J, Dobreva I, Maidan M, Gray V, et al. Rictor and integrin-linked kinase interact and regulate Akt phosphorylation and cancer cell survival. Cancer Res 2008; 68:1618 - 24; http://dx.doi.org/10.1158/0008-5472.CAN-07-5869; PMID: 18339839
- Gunst SJ, Zhang W. Actin cytoskeletal dynamics in smooth muscle: a new paradigm for the regulation of smooth muscle contraction. Am J Physiol Cell Physiol 2008; 295:C576 - 87; http://dx.doi.org/10.1152/ajpcell.00253.2008; PMID: 18596210
- Golovanov AP, Chuang TH, DerMardirossian C, Barsukov I, Hawkins D, Badii R, et al. Structure-activity relationships in flexible protein domains: regulation of rho GTPases by RhoGDI and D4 GDI. J Mol Biol 2001; 305:121 - 35; http://dx.doi.org/10.1006/jmbi.2000.4262; PMID: 11114252
- DerMardirossian C, Rocklin G, Seo JY, Bokoch GM. Phosphorylation of RhoGDI by Src regulates Rho GTPase binding and cytosol-membrane cycling. Mol Biol Cell 2006; 17:4760 - 8; http://dx.doi.org/10.1091/mbc.E06-06-0533; PMID: 16943322
- Wu Y, Moissoglu K, Wang H, Wang X, Frierson HF, Schwartz MA, et al. Src phosphorylation of RhoGDI2 regulates its metastasis suppressor function. Proc Natl Acad Sci U S A 2009; 106:5807 - 12; http://dx.doi.org/10.1073/pnas.0810094106; PMID: 19321744
- Dovas A, Choi Y, Yoneda A, Multhaupt HA, Kwon SH, Kang D, et al. Serine 34 phosphorylation of rho guanine dissociation inhibitor (RhoGDIalpha) links signaling from conventional protein kinase C to RhoGTPase in cell adhesion. J Biol Chem 2010; 285:23296 - 308; http://dx.doi.org/10.1074/jbc.M109.098129; PMID: 20472934
- Griner EM, Churchill ME, Brautigan DL, Theodorescu D. PKCα phosphorylation of RhoGDI2 at Ser31 disrupts interactions with Rac1 and decreases GDI activity. Oncogene 2012; In press http://dx.doi.org/10.1038/onc.2012.124; PMID: 22469974
- DerMardirossian C, Schnelzer A, Bokoch GM. Phosphorylation of RhoGDI by Pak1 mediates dissociation of Rac GTPase. Mol Cell 2004; 15:117 - 27; http://dx.doi.org/10.1016/j.molcel.2004.05.019; PMID: 15225553
- Yu J, Zhang D, Liu J, Li J, Yu Y, Wu XR, et al. RhoGDI SUMOylation at Lys-138 increases its binding activity to Rho GTPase and its inhibiting cancer cell motility. J Biol Chem 2012; 287:13752 - 60; http://dx.doi.org/10.1074/jbc.M111.337469; PMID: 22393046