Abstract
The main functional roles attributed to the centrosome, the major microtubule organizing center (MTOC) of metazoans, are related to cell locomotion, sensory perception and division. The role of vesicular trafficking in the regulation of the centrosome cycle has been largely unexplored. Recently, however, several studies have indicated the involvement of molecules and/or complexes of the trafficking routes in centrosome positioning, duplication and regulation. Functional screens have revealed communication between the outer nuclear envelope, the Golgi apparatus, the endosomal recycling compartment and centrosomes, while other studies underline the involvement of the ESCRT complex proteins in centrosome function. In this commentary, we discuss our recent study, which shows the involvement of an endosomal Rho protein, namely RhoD, in centrosome duplication and possible links between the centrosome’s structural and functional integrity to vesicular trafficking.
The Centrosome
The centrosome is the major microtubule-organizing center of animal cells, which influences cell shape and directs the formation of the bipolar mitotic spindle. At the core of a typical centrosome are two cylindrical microtubule-based structures termed centrioles, which recruit a matrix of associated pericentriolar material (PCM). Aberrations in numerical and structural integrity of centrosomes interfere with spindle formation and chromosome segregation. Centrosome duplication occurs through a conserved cycle.Citation1 It begins with centriole disengagement, followed by procentriole assembly perpendicular to the pre-existing centriole and occurs during the S phase of the cell cycle. By G2, both centriole pairs mature by recruiting necessary components such as PCM (maturation phase) and as cells proceed to M phase, the two centrosomes separate and orient toward opposing poles of the cells to drive the formation of the mitotic spindle. During separation, only one centrosome is functionally active (“dominant” or “mother” centrosome) in terms of microtubule nucleation and structural maturation and the other (“new” or “daughter” centrosome) moves to the opposite site and completes its maturation ~10 min before nuclear envelope breakdown.Citation2 Centrosome duplication is heavily regulated by cell cycle controls. The link between the cell and centrosome cycles is mediated through the function of cyclins and their cyclin-dependent kinases.
Centrosome positioning is crucial, as it determines the location of many other organelles within the cell and regulates proper cell division.Citation3 This localization is actively maintained via interconnected mechanical forces applied from microtubules and the cortical actin network. This crosstalk is essential not only for the maintenance of the central localization of the MTOC in interphase cells, but also for orchestrating the movement of the duplicated centrosomes during cell division ().Citation4-Citation6
Figure 1. Mechanical forces applied on centrosomes. Centrosome positioning is maintained by coordinated microtubule pulling forces and cortical actin pushing centripetal flow. During division, enhanced actomyosin contractility in the periphery of the opposite pole drags adjacent astral microtubules to drive the separation of duplicated centrosomes to opposing poles. Figure adapted from both Burakov and Rosenblatt.Citation4,Citation5 Centrosome (gray), nucleus (pink), recycling compartment (orange), endoplasmic reticulum (green), Golgi (magenta).
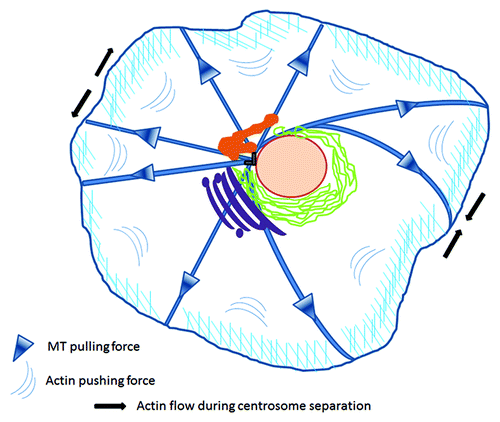
Changes in numbers, structure and/or location of centrosomes result in phenotypic abnormalities (from apoptosis to uncontrolled proliferation) due to chromosome instabilities. Centrosome amplification forces cells to pass through a multipolar spindle intermediate, enhancing merotelic attachments and lagging chromosomes, and as a consequence unequal chromosome segregation into the daughter cells.Citation7,Citation8 In fact, altered centrosomes have been described in many tumors.Citation9-Citation13 Furthermore, direct manipulation of centrosomes in vitro leads to aneuploidy and transformation, supporting a potentially causative effect toward malignancy.Citation14
Crosstalk Between Trafficking Routes and the Centrosome
Golgi apparatus
Over the last few years, a series of studies in centrosome research have re-evaluated the correlation of trafficking compartments and centrosome integrity. In interphase mammalian cells, the Golgi apparatus is found in close proximity to the centrosome and recent data suggest that this proximity is accompanied by a functional link between these two organelles. Golgi-associated proteins that control the pericentriolar position of the Golgi apparatus, also contribute to centrosome reorganization.Citation15,Citation16 Furthermore, GM130, a Golgi protein, by association with Tuba, a CDC42 guanine nucleotide exchange factor, activates CDC42, which in turn regulates centrosome organization.Citation17 The bidirectional interaction between the Golgi and centrosome is highlighted by the fact that centrosome-nucleated microtubules govern Golgi positioning.Citation18 In a recent study, functional repurposing revealed the involvement of the STRIPAK (Striatin-InteRacting Phosphatase and Kinase) signaling complexes in mediating communication between the outer nuclear envelope, the Golgi and centrosomes and is likely to unravel new mechanisms of organelle homeostasis.Citation19
ESCRT complex
Centrosome integrity is strongly affected by proteins involved in other trafficking routes as well. The endosomal sorting complex required for transport (ESCRT) is required for the recognition and sorting of ubiquitin-modified cargo proteins into the internal vesicles of multivesicular bodies (MVBs)Citation20-Citation22 and catalyzes membrane scission eventsCitation23 in the biogenesis of MVBs and cytokinesis.Citation24-Citation27 A recent study identified a functional role for late-acting ESCTIII/VPS4 proteins in centrosome and mitotic spindle maintenance.Citation28 Cells depleted of ESCRT-III/VPS4 exhibit highly penetrant phenotypes including multipolar spindles and over amplified centrosomes, suggesting a strong participation of the ESCRT complex in the proper centrosome cycle. Furthermore, ESCRT factors have also been shown to regulate spindle-pole-body duplication in the functional repurposing study mentioned above—further underlining the diversity of cellular pathways.Citation19
Recycling compartment
The perinuclear recycling endosomal compartment is in close proximity to the centrosomes, however, there is little information regarding possible functional or structural association between these organelles as yet. Recently, however, there are indications that such an association does indeed exist. The seminal study of Doxsey’s groupCitation29 identified a direct link between endosomes and centrosomes. More specifically, two mother centriole appendage proteins, centriolin and cenexin/ODF2, were found to regulate the association of the recycling endosome components Rab11-GTPase, the Rab11-GAP protein Evi5 and the exocyst complex at the mother centriole. According to the proposed model for the hierarchy of molecular interactions, cenexin anchors centriolin at the appendages of the mother centriole, which in turn recruits the exocyst complex, necessary for the recycling. The exocyst associates with Evi5 GAP, which inactivates Rab11, leading to the release of the GTPase from the centrosome. Interestingly, knockdown of centriolin resulted in an increase of recycling, suggesting that upon loss of centriolin and the binding partner Evi5 GAP from centrosomes, Rab11-GTP is retained on this structure enhancing the recycling of cargo from neighboring endosomes to the plasma membrane. These observations indicate a dual role of Rab11 both in endosomal function (efficient recycling) and in centrosome regulation as well.
Our group’s work provides an extra link. RhoD GTPase, an endosomal protein, belongs to the Rho family of GTPases and has been mainly implicated in endosome dynamics. More specifically overexpression of a GTPase deficient mutant of RhoD (RhoDG26V) impedes the motility of early endosomes, which are scattered in the periphery of the cell.Citation30 A novel splice variant of human Diaphanous, hDia2C, mediates the effect of RhoD on endosome motility and involves the membrane recruitment and activation of c-Src.Citation31 Our current work describes a novel function of RhoD potentially connecting membrane trafficking and the centrosome cycle.Citation32 In this paper we describe novel effects of RhoD in terms of both cell and centrosome cycle progression. Our findings suggest that RhoD promotes G1/S transition via the interaction with a newly identified effector, Diaphanous1 and that dominant activation or depletion of RhoD interferes with centrosome integrity. Expression of RhoDG26V results in atypical nuclear morphology seen both in vivo and in vitro, characterized by the formation of cells with nuclear fragmentation and multinucleation. Further analysis revealed that nuclear fragmentation was associated with chromosome misalignment and formation of multipolar spindles during metaphase, whereas cells processing more than one “normal” sized nucleus were generated upon cytokinesis defects. This effect is so striking that the cells are lost within a few days due to apoptosis.
RhoDG26V promotes centriole overduplication, whereas expression of the dominant negative RhoDT31N or knockdown of the protein abrogates centrosome duplication. Electron microscopic analysis revealed the existence of cells with atypical centrosomal structures or even cells lacking centrosomal structures completely. This is a striking example of an endosomal protein interfering structurally with the centrosome to the extent that the lack of this protein inhibits the formation of the centrosome. Detailed imaging indicates that RhoD is localized in the pericentriolar area in G1 phase and as the cell progresses to S-G2 phase, RhoD positive structures “decorate” one of the two centrosomes, which preliminary data hint is the mother centriole ( and with PCM). In conjunction with these data, super-resolution confocal imaging reveal an important overlap with Rab11 positive pericentriolar structures (, unpublished data; ), underlying the importance of RhoD in the regulation of the centrosome cycle. What is the mechanism of action of RhoD in centrosome duplication? It is likely that alterations of the actin cytoskeleton contribute to the effect of RhoD, however trafficking may also be involved and the delivery of components required for centrosome assembly may be altered. We are currently addressing these issues.
Figure 2. RhoD encircles centrosomes. Preference to one of the two centrin positive couples. HUVE cells were infected with myc-RhoDWT or myc-RhoDG26V and transfected with GFP-Centrin. Twenty four-hours post transfection/infection cells were processed for GFP-centrin, myc-RhoDWT or G26V (red-TRITC) and endogenous α-tubulin (blue-CY5). Bar = 10 μm. Image taken from Kyrkou.Citation32
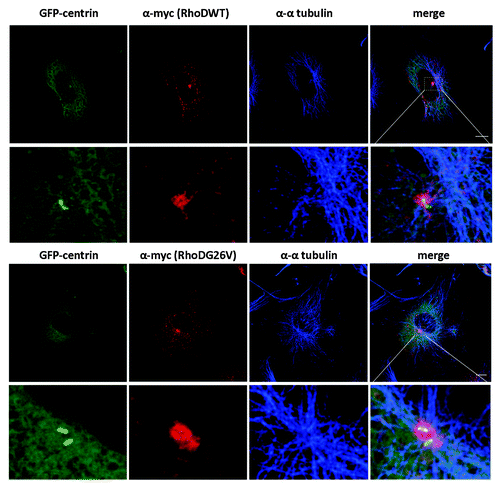
Figure 3. RhoDWT does not colocalize with PCM1-positive centrosome granules. HUVE cells were transfected with myc-RhoDWT for 24 h. Myc-RhoDWT protein was detected with α-myc/Alexa α-mouse 488 and centrosome granules via α-PCM1/ATTO α-rabbit 425. The upper panel is a confocal image. The boxed region in the upper image is shown in the lower panel and is a STED image. Bar = 10 μm.
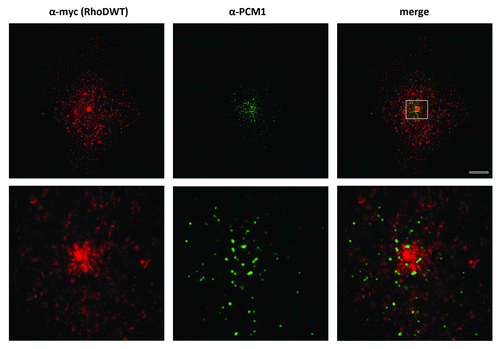
Figure 4. RhoDWT-positive endosomes show partial overlap with Rab11-positive (recycling) endosomes. HUVE cells were co-transfected with myc-RhoDWT and GFP-Rab11WT for 24 h. GFP fluorescence was quenched by fixation in methanol for 20 min at −20°C. Myc-RhoD was detected with α-myc/ATTO-rabbit 425 and Rab11 with α-GFP/Alexa α-mouse 488. The upper panel is a confocal image. The boxed regions in the upper merged image are shown in the central and lower panels and are STED images. The central panels show the colocalization of Rab11 and RhoD in the perinuclear recycling compartment, while the lower panels show the colocalization on peripheral vesicles. Bar = 5 μm. Quantitation with MotionTracking Software yield approximately 56% overlap of RhoD-positive vesicles vs. Rab11-positive endosomes (and 32% of Rab11/RhoD). This is based on total colocalization, while in the perinuclear recycling compartment the colocalization is significantly higher.
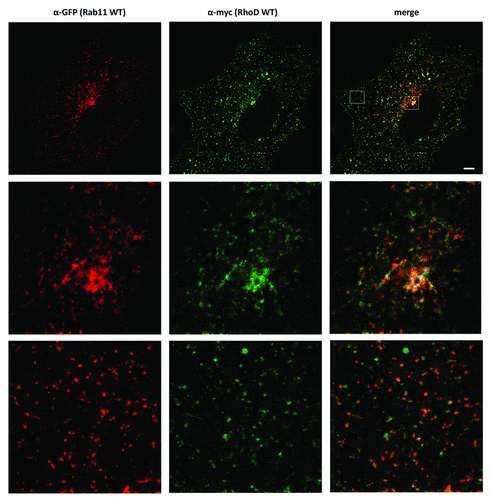
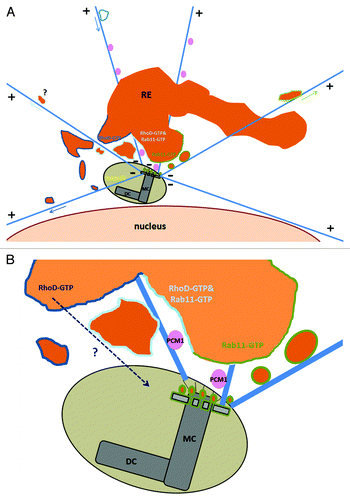
Figure 5. Crosstalk between the recycling compartment and the centrosome. (A) RhoD-positive and/or Rab11 positive vesicles of the recycling compartment surround the centrosome. A distinct population of endosomes (Microtubule driven trafficking in arrow) are either moving from the recycling area (or maybe the periphery) toward the centrosome or emanating from this pericentriolar area to the cell’s periphery. (B) Mother centriole appendages are decorated with recycling endosomes and membrane-free Rab11-GTP.Citation29 RhoD-GTP trafficking inside the centrosome structure is under investigation. Illustrated areas of RhoD-GTP in dark blue lines, Rab11-GTP in green lines, RhoD-GTP and Rab11-GTP colocalization in cyan, PCM1 granules in pink, Rab6c in yellow. MC, mother centriole; DC, daughter centriole.
Implication of Other GTPases in the Centrosome Cycle Regulation
To date few GTPase have been implicated in centrosome integrity. Another member of the Rab family, Rab6c has been recently identified as a regulator of the centrosome cycle.Citation33 Rab6c is a retrogene, transcribed in a limited number of human tissues including brain, testis, prostate and breast and depletion of this isoform generates tetraploid cells with supernumerary centrosomes, via an as yet unidentified mechanism. On the other hand, the Rho family members participate in a more dynamic way in the centrosome cycle since both RhoA and RhoC (but not RhoB) are related to the amplification phase by activating ROCKII.Citation34 Activated mutants of RhoA/C promote centriole amplification whereas knockdown-interfere with the formation of a functional structure. Interestingly, depletion of RhoA is compensated by the expression of RhoC and vice versa, implying that it is possible that RhoA/C may comprise the intracellular concentration of total Rho proteins required for the initiation of centrosome duplication.
Centrioles also form the basal bodies required for the formation of cilia and flagella. Cilia lack protein synthesis and therefore their formation relies on protein trafficking. Toward this purpose, both the conserved intraflagellar transport system and other transport regulators including Rab8, Rabin8, Arf4, Rab11 and the exocyst complex are used.Citation35,Citation36 Recently regulators of endocytosis have also been implicated in ciliary membrane trafficking.Citation37 As cilia are nucleated from centrioles it has been recently shown that extra centrioles and cilia disrupt ciliary signaling.Citation38 Therefore, as RhoD regulates centriole duplication and is an endosomal protein regulating endosome motility, it is likely that RhoD may also affect ciliary function, this is under investigation.
Conclusion
Overall, it seems that the functional significance of the physical proximity of Golgi and recycling compartments to centrosomes is becoming clear. Identification of complexes mediating communication between these organelles suggests reciprocal homeostasis mechanisms. It is likely that centrosome function will be influenced by components of the recycling endosomes and Golgi having possible implications in asymmetric cell division and ciliary signaling. Further investigation is required to gain insight into the molecular mechanisms underlying this cooperation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Meraldi P, Nigg EA. The centrosome cycle. FEBS Lett 2002; 521:9 - 13; http://dx.doi.org/10.1016/S0014-5793(02)02865-X; PMID: 12067716
- Rusan NM, Peifer M. A role for a novel centrosome cycle in asymmetric cell division. J Cell Biol 2007; 177:13 - 20; http://dx.doi.org/10.1083/jcb.200612140; PMID: 17403931
- Manneville JB, Etienne-Manneville S. Positioning centrosomes and spindle poles: looking at the periphery to find the centre. Biol Cell 2006; 98:557 - 65; http://dx.doi.org/10.1042/BC20060017; PMID: 16907664
- Burakov A, Nadezhdina E, Slepchenko B, Rodionov V. Centrosome positioning in interphase cells. J Cell Biol 2003; 162:963 - 9; http://dx.doi.org/10.1083/jcb.200305082; PMID: 12975343
- Rosenblatt J, Cramer LP, Baum B, McGee KM. Myosin II-dependent cortical movement is required for centrosome separation and positioning during mitotic spindle assembly. Cell 2004; 117:361 - 72; http://dx.doi.org/10.1016/S0092-8674(04)00341-1; PMID: 15109496
- Cao J, Crest J, Fasulo B, Sullivan W. Cortical actin dynamics facilitate early-stage centrosome separation. Curr Biol 2010; 20:770 - 6; http://dx.doi.org/10.1016/j.cub.2010.02.060; PMID: 20409712
- Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature 2009; 460:278 - 82; http://dx.doi.org/10.1038/nature08136; PMID: 19506557
- Quintyne NJ, Reing JE, Hoffelder DR, Gollin SM, Saunders WS. Spindle multipolarity is prevented by centrosomal clustering. Science 2005; 307:127 - 9; http://dx.doi.org/10.1126/science.1104905; PMID: 15637283
- Duensing S, Münger K. Centrosome abnormalities, genomic instability and carcinogenic progression. Biochim Biophys Acta 2001; 1471:M81 - 8; PMID: 11342187
- Gisselsson D, Jonson T, Yu C, Martins C, Mandahl N, Wiegant J, et al. Centrosomal abnormalities, multipolar mitoses, and chromosomal instability in head and neck tumours with dysfunctional telomeres. Br J Cancer 2002; 87:202 - 7; http://dx.doi.org/10.1038/sj.bjc.6600438; PMID: 12107843
- D’Assoro AB, Lingle WL, Salisbury JL. Centrosome amplification and the development of cancer. Oncogene 2002; 21:6146 - 53; http://dx.doi.org/10.1038/sj.onc.1205772; PMID: 12214243
- Lingle WL, Barrett SL, Negron VC, D’Assoro AB, Boeneman K, Liu W, et al. Centrosome amplification drives chromosomal instability in breast tumor development. Proc Natl Acad Sci U S A 2002; 99:1978 - 83; http://dx.doi.org/10.1073/pnas.032479999; PMID: 11830638
- Mayer F, Stoop H, Sen S, Bokemeyer C, Oosterhuis JW, Looijenga LH. Aneuploidy of human testicular germ cell tumors is associated with amplification of centrosomes. Oncogene 2003; 22:3859 - 66; http://dx.doi.org/10.1038/sj.onc.1206469; PMID: 12813459
- Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, et al. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet 1998; 20:189 - 93; http://dx.doi.org/10.1038/2496; PMID: 9771714
- Bisel B, Wang Y, Wei JH, Xiang Y, Tang D, Miron-Mendoza M, et al. ERK regulates Golgi and centrosome orientation towards the leading edge through GRASP65. J Cell Biol 2008; 182:837 - 43; http://dx.doi.org/10.1083/jcb.200805045; PMID: 18762583
- Kodani A, Sütterlin C. The Golgi protein GM130 regulates centrosome morphology and function. Mol Biol Cell 2008; 19:745 - 53; http://dx.doi.org/10.1091/mbc.E07-08-0847; PMID: 18045989
- Kodani A, Kristensen I, Huang L, Sütterlin C. GM130-dependent control of Cdc42 activity at the Golgi regulates centrosome organization. Mol Biol Cell 2009; 20:1192 - 200; http://dx.doi.org/10.1091/mbc.E08-08-0834; PMID: 19109421
- Cole NB, Sciaky N, Marotta A, Song J, Lippincott-Schwartz J. Golgi dispersal during microtubule disruption: regeneration of Golgi stacks at peripheral endoplasmic reticulum exit sites. Mol Biol Cell 1996; 7:631 - 50; PMID: 8730104
- Frost A, Elgort MG, Brandman O, Ives C, Collins SR, Miller-Vedam L, et al. Functional repurposing revealed by comparing S. pombe and S. cerevisiae genetic interactions. Cell 2012; 149:1339 - 52; http://dx.doi.org/10.1016/j.cell.2012.04.028; PMID: 22682253
- Gruenberg J, Stenmark H. The biogenesis of multivesicular endosomes. Nat Rev Mol Cell Biol 2004; 5:317 - 23; http://dx.doi.org/10.1038/nrm1360; PMID: 15071556
- Hurley JH. ESCRT complexes and the biogenesis of multivesicular bodies. Curr Opin Cell Biol 2008; 20:4 - 11; http://dx.doi.org/10.1016/j.ceb.2007.12.002; PMID: 18222686
- Piper RC, Katzmann DJ. Biogenesis and function of multivesicular bodies. Annu Rev Cell Dev Biol 2007; 23:519 - 47; http://dx.doi.org/10.1146/annurev.cellbio.23.090506.123319; PMID: 17506697
- Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature 2009; 458:172 - 7; http://dx.doi.org/10.1038/nature07836; PMID: 19234443
- Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science 2007; 316:1908 - 12; http://dx.doi.org/10.1126/science.1143422; PMID: 17556548
- Lindås AC, Karlsson EA, Lindgren MT, Ettema TJ, Bernander R. A unique cell division machinery in the Archaea. Proc Natl Acad Sci U S A 2008; 105:18942 - 6; http://dx.doi.org/10.1073/pnas.0809467105; PMID: 18987308
- Morita E, Sandrin V, Chung HY, Morham SG, Gygi SP, Rodesch CK, et al. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J 2007; 26:4215 - 27; http://dx.doi.org/10.1038/sj.emboj.7601850; PMID: 17853893
- Samson RY, Obita T, Freund SM, Williams RL, Bell SD. A role for the ESCRT system in cell division in archaea. Science 2008; 322:1710 - 3; http://dx.doi.org/10.1126/science.1165322; PMID: 19008417
- Morita E, Colf LA, Karren MA, Sandrin V, Rodesch CK, Sundquist WI. Human ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance. Proc Natl Acad Sci U S A 2010; 107:12889 - 94; http://dx.doi.org/10.1073/pnas.1005938107; PMID: 20616062
- Hehnly H, Chen CT, Powers CM, Liu HL, Doxsey S. The centrosome regulates the Rab11- dependent recycling endosome pathway at appendages of the mother centriole. Curr Biol 2012; 22:1944 - 50; http://dx.doi.org/10.1016/j.cub.2012.08.022; PMID: 22981775
- Murphy C, Saffrich R, Grummt M, Gournier H, Rybin V, Rubino M, et al. Endosome dynamics regulated by a Rho protein. Nature 1996; 384:427 - 32; http://dx.doi.org/10.1038/384427a0; PMID: 8945468
- Gasman S, Kalaidzidis Y, Zerial M. RhoD regulates endosome dynamics through Diaphanous-related Formin and Src tyrosine kinase. Nat Cell Biol 2003; 5:195 - 204; http://dx.doi.org/10.1038/ncb935; PMID: 12577064
- Kyrkou A, Soufi M, Bahtz R, Ferguson C, Bai M, Parton RG, et al. RhoD participates in the regulation of cell-cycle progression and centrosome duplication. Oncogene 2012; http://dx.doi.org/10.1038/onc.2012.195; PMID: 22665057
- Young J, Ménétrey J, Goud B. RAB6C is a retrogene that encodes a centrosomal protein involved in cell cycle progression. J Mol Biol 2010; 397:69 - 88; http://dx.doi.org/10.1016/j.jmb.2010.01.009; PMID: 20064528
- Kanai M, Crowe MS, Zheng Y, Vande Woude GF, Fukasawa K. RhoA and RhoC are both required for the ROCK II-dependent promotion of centrosome duplication. Oncogene 2010; 29:6040 - 50; http://dx.doi.org/10.1038/onc.2010.328; PMID: 20697357
- Deretic D, Wang J. Molecular assemblies that control rhodopsin transport to the cilia. Vision Res 2012; 75:5 - 10; http://dx.doi.org/10.1016/j.visres.2012.07.015; PMID: 22892112
- Drummond IA. Cilia functions in development. Curr Opin Cell Biol 2012; 24:24 - 30; http://dx.doi.org/10.1016/j.ceb.2011.12.007; PMID: 22226236
- Kaplan OI, Doroquez DB, Cevik S, Bowie RV, Clarke L, Sanders AA, et al. Endocytosis genes facilitate protein and membrane transport in C. elegans sensory cilia. Curr Biol 2012; 22:451 - 60; http://dx.doi.org/10.1016/j.cub.2012.01.060; PMID: 22342749
- Mahjoub MR, Stearns T. Supernumerary centrosomes nucleate extra cilia and compromise primary cilium signaling. Curr Biol 2012; 22:1628 - 34; http://dx.doi.org/10.1016/j.cub.2012.06.057; PMID: 22840514