Abstract
Guanine nucleotide exchange factors (GEFs) control the site and extent of GTPase activity. Longin domains (LDs) are found in many Rab-GEFs, including DENNs, MON1/CCZ1, BLOC-3 and the TRAPP complex. Other GEFs, including Ragulator, contain roadblock domains (RDs), the structure of which is closely related to LDs. Other GTPase regulators, including mglB, SRX and Rags, use LDs or RDs as platforms for GTPases. Here, we review the conserved relationship between GTPases and LD/RDs, showing how LD/RD dimers act as adaptable platforms for GTPases. To extend our knowledge of GEFs, we used a highly sensitive sequence alignment tool to predict the existence of new LD/RDs. We discovered two yeast Ragulator subunits, and also a new LD in TRAPPC10 that may explain the Rab11-GEF activity ascribed to TRAPP-II.
Introduction
Every small GTPase is turned on by a guanine nucleotide exchange factor (GEF), and inactivated by a GTPase activation protein (GAP). Despite the large number of GEFs now known, many GTPases have no identified GEF, indicating that more remain to be discovered. For example, there are 66 Rabs that regulate membrane traffic in humans, with GEFs identified for only 75% of these.Citation1-Citation3 A recurrent theme among Rab-GEFs in particular, but other GEFs as well, is the presence of a longin domain (LD), and a careful analysis of LDs and their complexes indicates some useful rules.
LDs are α-β-α sandwiches of approximately 100–120 aa consisting of a 5 sheet β-meander (⇑⇓⇑⇓⇑ pattern) with one helix on one side (which we arbitrarily define as the top), and two helices on the other side (bottom). RDs, described in more detail in the next section, are almost identical. LDs and RDs are protein interaction modules, but the range of their interactions is highly restricted. The majority of complexes that contain LDs or RDs also contain GTPases (). The exceptions to this include the LDs found as N-terminal extensions of a sub-family of SNAREs called longins because they are the longest SNAREs. Here the LDs fold back to bind the SNARE helix.Citation4,Citation5 Also, small and medium subunits of major coat complexes contain LDs, the function of which is generally unknown, although one binds Arf1.Citation6 Finally, DYNLRB1 is an RD-only Dynein Light Chain-7 that has many different binding partners, one of which is a Rab-GTPase.Citation7
Figure 1. Complexes with longin or roadblock domains that are proposed to interact with small GTPases. Diagram of 8 complexes that contain small GTPases and longin domains (LD) or roadblock domains (RD), with associated Protein Data Bank (PDB) codes if structures have been solved:- mglB dimer: mglA (3T1Q); SRα: SRβ (2FH5); Rag C-termini: Rag N-termini (4ARZ); DENND1B: Rab35 (3TW8); MON1/CCZ1: Rab7; NPRL2/3: GTPase not known; Ragulator, which contains two dimers: MP1/p14 (1VET, 4FTX) and HBXIP/C7orf59: Rag-GTPase dimer (specific relationship not known); TRAPP-C1/C4: Rab1 (3CUE). Domains of known and predicted structure are colored as follows: LDs and RDs, light blue with dimerization interfaces in dark blue, GTPases, red; DENN C-terminus, green; helix-turn-helix, orange. Domains of unknown structure (yellow) include: MON1 C-terminus, CCZ1 C-terminus and the fifth subunit of Ragulator, p18/Ego1p. Black lines link domains in multidomain proteins. For each complex, the effect of the LD/RD on GTPase activation is indicated: + = GEF; (-) = GAP (indirectly); 0 = neither GEF nor GAP; ? = not known. For TRAPP, the LD dimer is surrounded by multiple other subunits (indicated by dashed lines), described in detail in .
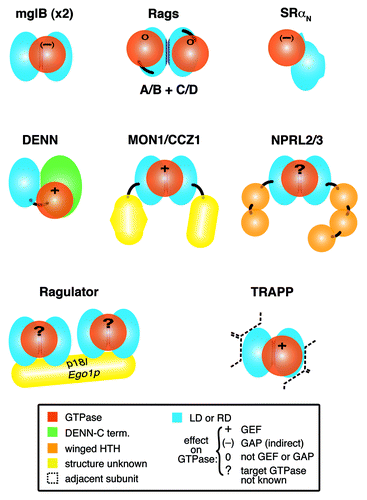
Here we have surveyed recent advances in our knowledge of the solved and predicted structures of complexes containing LD/RDs, leading to the general rule that pairs of LD/RDs dimerize to form a platform for a GTPase. We also discovered distant homologs of LD/RDs in two cases, finding a new pair of RD subunits for Ragulator in yeast, and a new LD in the transport protein particle (TRAPP)-II complex. This LD in TRAPP-II completes a pair of LDs, and may therefore contribute to a platform for an extra GTPase.
Current Knowledge
MglA/B: Roadblock domains as the ancestors of Longin domains
Most proteins with LDs are conserved in metazoal, fungal and plant homologs,Citation8 indicating that LD diversification was present in the last eukaryotic common ancestor. Looking at more distantly related cells, there are no LDs in prokaryotes. However, a simple circular permutation switches the last element of an LD (ββαβββαα) to become the first element of a Roadblock domain (RD) (αββαβββα) (), and RDs are found in complexes with small GTPases in bacteria and archea as well as in eukaryotes.Citation7,Citation9,Citation10 Thus, LD/RD-GTPase complexes are ubiquitous, and RDs are good candidates for the ancestral domain from which LDs developed. Can anything be said about the arrangement of subunits in the ancient complex?
Figure 2. Longin and Roadblock domains form highly conserved platforms for small GTPases. (A) Diagram of a symmetric LD dimer. Structural elements in an LD are: a five-stranded β-meander oriented in the plane of the page (blue); the top face with a single helix (green), and the bottom face with two helices (yellow). RDs are circularly permuted forms with alternate start/stop sites (asterisks). Labels and connectors for the left-hand LD are in white and gray, and for the right-hand LD in black. The dimer interface consists of β3 and α1. A conformational switch in α1 from a short form (15 residues, 4 turns) to a long form (27 residues, 6.5 turns, extension labeled α1’) is found in some LDs, for example TRAPPC2 and TRAPPC4, and is associated with dimerization.Citation46,Citation54 (B) A homodimer of bacterial mglB forms an RD platform for the small GTPase mglA. The extent of the GTPase binding site is indicated by the red circle.Citation10 Color scheme as in A., with transparent green helices on the upper face. The RD only proteins MP1 and p14 form an almost identical dimer in human Ragulator.Citation9 (C) Heterodimers in TRAPP-I form an LD platform for Rab1. The complex of TRAPPC4 (also called synbindin, in yeast Trs23p), TRAPPC1 (Bet5p) and Rab1 (Ypt1p) is highly similar to mglA/B.Citation12 (D) DENN forms a platform for Rabs that includes an LD. DENND1B has a bi-lobed structure, with an N-terminal LD (residues 1-149, colors as in A) and a C-terminal α-β domain (residues 170–410, gray). Rab35 binds to the top surface, but considerably off center. A minority (6/21) of contacts is predicted to be with the LD.Citation13 (E) Different organization of the RD dimer in yeast Ego3p. Compared with parts A-D (above), the dimer is shown rotated 180° (upside-down). α1 (equivalent to α3 in LDs) is swapped between domains. The top helix (α2, in LDs = α1) does not approach the dimer interface, which consists of a sixth sheet adjacent to β3 (*), and the bottom helix (α3, in LDs = α2).Citation16 Accession numbers in parts B-E: 3T12, 3CUE, 3TW8 and 4FTX.
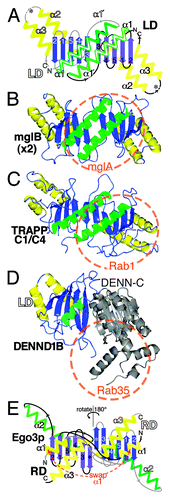
The sole bacterial RD with a solved structure is mglB, an RD-only protein that binds mglA, a GTPase that regulates cell gliding ().Citation10 MglB homo-dimerizes as a result of two intermolecular interactions: (1) β-sheet dimerization by β3, forming a ten-stranded β-meander; (2) interactions between the side chains of the helices on the top surface that form the binding site for GTPase ().Citation10 MglB facilitates reorientation within mglA so that an arginine (Arg53) intrinsic to the GTP-binding domain participates in the GTP-hydrolysis reaction.Citation10 Thus, mglB is not catalytically active, though it facilitates GAP activity.
The indirect way in which mglB affects the GDP/GTP state of mglA is typical of how LDs and RDs interact with GTPases in general. They are not active catalysts that directly interact with nucleotide, instead they are platforms for GTPases, often with extra subunits or domains recruited either as GAP,Citation10,Citation11 or (more typically) as GEF.Citation12,Citation13 Another aspect of the mglB-mglA complex that is shared with other LD/RD complexes, is that it contains an LD/RD dimer and has a binding site for GTPase binding on its top surface (the surface containing a single helix from each LD or RD). This relationship applies to LD/RDs throughout evolution ().Citation12-Citation16 However, as with many rules, there are exceptions: the Signal recognition particle Receptor (SR) α subunit contains a monomeric LD adapted so that it cannot dimerize, although a GTPase still binds to its top surface ().Citation11
Rag-GTPases: Intrinsic RDs as a platform for GTPases without regulation of GTP:GDP binding
Target of rapamycin (TOR) kinases form two conserved complexes TORC1 and TORC2 that regulate many cellular functions in response to nutritional status. TORC1 responds to cellular levels of amino acids after recruitment to lysosomal membranes by raptor, a TORC1 component that in its turn is recruited by a dimer of Rag-GTPases.Citation17 In this dimer either RagA or RagB binds to one of RagC or RagD, and the most active conformation is RagA/B-GTP plus RagC/D-GDP (equivalent to Gtr1p-GTP plus Gtr2p-GDP in yeast).Citation15,Citation17 The four Rag-GTPases differ from all other small GTPases by having an intrinsic RD in cis, C-terminal to the GTPase (). The RDs mediate Rag dimerization, and the crystal structure of the RDs is very similar to mglA/B, with the GTPases binding to the helices on the top surface ().Citation14,Citation16 The interaction sites for the two Rag GTPase domains are separated, and they can switch between active and inactive conformations independently.Citation15,Citation16 There is one difference between Rags and other known LD/RD-GTPase complexes: the Rag-GTPase domains are rotated so that the GTP pockets are far away from the RD platform.Citation14,Citation15 This rotation may have evolved here because the GTPase domains interact not just with these RDs, but also with RDs in the Ragulator complex (see below).
DENNs: An expanding superfamily of GEFs
The largest family of Rab-GEFs are the DENNs (standing for differentially expressed in normal cells and neoplasia). There are 17 DENNs in human, all sharing a domain of 400-500 aa.Citation1,Citation18 The domain sub-structure was previously defined as a three-way division into u-DENN/c-DENN/d-DENN “domains,” but a the solved structure of DENND1B together with Rab35 has made this nomenclature obsolete. The structure consists of two intimately associated lobes.Citation13 The N-terminal lobe is an LD, and the C-terminal lobe is a complex α-β fold with no known relatives. Thus, DENNs resemble half of an LD platform where one of the LDs has been replaced with an alternate partner ( and ). Rab35 interacts mainly with the DENN C-terminal lobe, and also with α1 and the β3-β4 loop of the LD. This results in a conformational change in the switch I and II regions of Rab35, displacing the conserved Phe35 residue from the nucleotide-binding pocket. These changes reduce the affinity for GDP, which is normally stabilized by interactions with the aromatic side chain of Phe35.Citation13
The DENN superfamily was recently expanded when the structure of the C-terminus of folliculin (FLCN) was found to be similar to the C-terminal lobe of DENN; in addition this portion of FLCN had Rab-GEF activity in vitro.Citation19 FLCN is a tumor suppressor that is mutated in Birt-Hogg-Dubé syndrome, leading to renal cancer and other tumors, partly through deregulation of TOR kinase.Citation20 The physiological target of FLCN is not known, but it may act on TOR indirectly via a Rab that regulates membrane traffic of components of TORC1 or TORC2.Citation21
The activity of DENN-like proteins tends to be restricted to particular target Rab GTPases, though there is a single DENN-like protein in yeast that binds an Arf GTPase.Citation22 While DENND1B is specific for Rab35, other DENNs are only weakly active toward this Rab.Citation13 DENND2-DENND5 display specific GEF activity toward other Rab GTPases.Citation1 Despite this restricted in vivo activity, when DENNs are presented with a range of Rabs in vitro they can nevertheless show weak non-specific GEF activity. For example, the C-terminal portion of FLCN shows some activity as a Rab35-GEF, even though Rab35 is not the physiological substrate.Citation19 In another example, the fruit fly DENND4 ortholog CRAG, which specifically activates Rab10 in fly and human cells to regulate baso-lateral secretion,Citation1,Citation23,Citation24 has weak activity toward Rab11. CRAG stimulated nucleotide exchange 1.25-fold in Rab11 compared with 35-fold in Rab10, its likely physiological substrate (see Fig. 6 in ref. Citation24). This shows that GEF activities must be interpreted with great care.
The finding that FLCN is like DENN, while sharing no obvious sequence homology by PSI-BLAST, indicates that the DENN superfamily is larger than first thought. There are two groups of proteins that can be added into the DENN superfamily. One group are the proteins with slight sequence homology to DENN or FLCN; these are likely to be Rab-GEFs. This applies to AVL9, LCHN, FAM45 (similar to DENN) and to Smith-Magenis candidate region 8 protein (similar to FLCN). The second group are proteins with similar secondary structure to DENN or FLCN, which are also likely to be share function. Using a sensitive alignment tool that includes secondary structural predictions,Citation25 we and others have found two new sets of DENN-like proteins: (1) FLCN-interacting proteins (FNIPs); and (2) the protein encoded by C9orf72, which we named DENN-like-72 (DENNL72) on the strength of its full-length homology to DENNs.Citation26,Citation27 Each of these families raises interesting points. FNIPs form complexes with FLCN, indicating that two DENN-like proteins can be present in the same complex. This suggests novel (and testable) modes of interaction, including simultaneous engagement of two GTPases. C9orf72/DENNL72 is interesting because it is the locus of the single most common mutation in familial fronto-temporal dementia and amyotrophic lateral sclerosis (motor neuron disease), and a major locus for sporadic forms of these diseases.Citation28 The mutation, which is dominant, is a 6.5–12 kb expansion of a hexanucleotide repeat in a non-coding intron. Our results suggest that in addition to gain of function effects of these repeats, DENNL72 haplo-insufficency might critically reduce Rab activation, inhibiting an aspect of endocytosis or autophagocytosis needed for neuronal survival.
MON1/CCZ1: Founding members of a group of Rab-GEFs with paired LDs
Another GEF that contains LDs is MON1/CCZ1, where both MON1 and CCZ1 have LDs,Citation29 and the complex is a Rab7-GEF.Citation30 A similar complex is formed by HPS1 and HPS4, homologs of MON1 and CCZ1 respectively. HPS1 and HPS4 form the BLOC-3 complex, which we recently identified as a Rab32/38-GEF.Citation2 In addition to sequence relationships between MON1/CCZ1 and BLOC-3, the complexes are functionally related, in that both complexes act on late endocytic compartments, and Rab7 is phylogenetically related to Rab32 and Rab38.Citation3 Since the components of BLOC-3 dimerize in part through their LDs,Citation31 it is possible, though not yet tested experimentally, that the LDs of both complexes form β-sheet dimers (). The GEF mechanism is also not yet known, but analogous to DENNs, we consider it likely that one or both C-terminal regions make key contacts that activate GDP:GTP exchange.
NPRL2/NPRL3 complexes as candidates for a novel Rab GEF regulating TORC1
Searches for distant homology have not only identified new full-length homologs of DENNs, but also new proteins with an LD, which by itself suggests interaction with a GTPase. LDs were found in two proteins that form a complex: nitrogen permease-like-2 (NPRL2), and its partner (and homolog) NPRL3.Citation26,Citation27 The LDs are at the N-termini of these proteins, and their C-termini contain two or more repeated ααααββ winged helix turn helix (HTH) domains (; Fig. S1).Citation32 Winged HTH repeats might bind DNA, RNA or protein, and in this case there is some evidence for a DNA partner, because one fungal NPRL2 homolog has a C-terminal extension containing an Exonuclease III domain.
In yeast, amino acid starvation activates Npr2p/Npr3p (the homologs of NPRL2/NPRL3) to inhibit TORC1,Citation33 possibly by direct interaction with TORC1 on either the vacuole or on phagophore assembly sites, where Npr2p/Npr3p complexes have different sets of binding partners,Citation34,Citation35 In humans NPRL2 and NPRL3 also dimerize,Citation33 though it is yet to be tested if this is through β-sheet LD dimerization. Could the LDs act as a platform for a GTPase involved in TORC1 signaling? Since RagC/D deliver a negative signal directly to TORC1,Citation15,Citation17 one possibility is that NPRL2/NPRL3 is a RagC/D-GEF.
New Discoveries
Ragulator: Prediction of a new pair of RDs in yeast Ragulator homologous to HBXIP and C7orf59
Ragulator is a complex that activates the Rag GTPase heterodimer.Citation36 There are five subunits in mammalian Ragulator, all small (14–20 kDa) single domain proteins, four of which are RD-only proteins that form two dimers ().Citation37 MP1 and p14 form an RD dimer with a structure highly similar to mglB.Citation9,Citation36 C7orf59 and HBXIP are recently described Ragulator subunits, and bind each other, so may also form a β-sheet dimer.Citation37 The fifth Ragulator subunit, p18, anchors the other subunits on lysosomes. It is possible that each RD heterodimer interacts with one Rag, although no functional studies have been reported. The Ragulator complex is conserved in yeast as the Ego complex (also called the Gse complex), which consists of Ego1p (also called Meh1p and Gse2p) that is distantly homologous to p18, and the RD-only protein Ego3p (also called Slm4p and Gse1p) that forms homodimers equivalent to the MP1/p14 heterodimer.Citation16,Citation38 Ego3p dimers swap the α1 helices on the bottom surface of the RDs, which involves a long unstructured loop between α1 and β1 ().Citation16 This radically alters the dimer interface even though the overall α-β-α sandwich is conserved.
Current analysis has not identified yeast counterparts of C7orf59 or HBXIP.Citation37 Since it is possible that small RD-only proteins are hard to find with standard techniques such as PSI-BLAST, we searched for distant structural yeast homologs of these human Ragulator subunits. We found that HBXIP and C7orf59 are both homologous to a pair of closely related small yeast proteins Ycr075wc-ap and Ynr034w-ap (). Because of their small size, these have been largely overlooked, and their interactions are poorly studied compared with other yeast proteins. However, the structure of Ynr034w-ap has been solved, and it adopts an RD-like fold, strongly supporting the evolutionary link we have uncovered ().Citation39 The structure of Ynr034w-ap (and that predicted for HBXIP and C7orf59) is αββαβββ, which is RD-like but missing α3. Ycr075w-ap is even shorter (ββαβββ), lacking both helices on the bottom surface. This is similar to DYNLRB1, which lacks α3, but where the helix is provided by its binding partner.Citation40 It is possible that the helices missing from Ragulator RDs are provided by p18/Ego1p, which is predicted to be largely helical (data not shown).
Figure 3. Structural homologies between known Ragulator subunits and new RD proteins in yeast. (A) Pair-wise matches obtained in HHpred searches seeded with MP1, p14, HBXIP, C7orf59, Ego3p, Ycr075w-ap and Ynr034w-ap (number of residues in brackets). The probabilities of shared structure (pss) obtained using HHpred are shown, together with the number of aligned residues (in brackets), and shading to indicate extent of pss, as previously.Citation27 Asterisks indicate where hits to proteins with solved structures in PDB (MP1/P14: 1VET; HBXIP: 3MSH; Ego3p: 4FTX; and Ynr034w-ap: 2GRG) were stronger than to the proteins and their structures predicted by HHpred. Yellow boxes show that HBXIP and C7orf59 are related to Ycr075w-ap and Ynr034w-ap. The Ycr075w-ap and Ynr034w-ap proteins are highly related to each other (orange boxes), with primary sequence homology detectable by PSI-BLAST (2nd iteration). pss = 100% for self-searches. (B) The NMR structure of Ynr034w-ap, available in PDB as 2GRG. In the RD-like fold, helix α3 is replaced by a segment of 15 highly mobile residues (white, other colors as ).Citation39
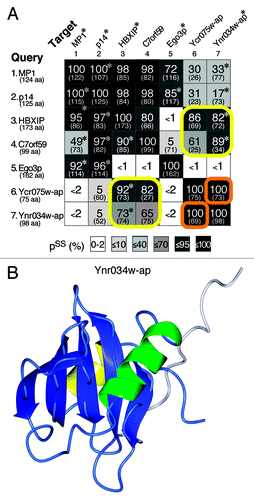
The close relationship between Ycr075w-ap and Ynr034w-ap suggests the possibility that they bind each other, as do C7orf59 and HBXIP.Citation37 Looking at other relationships, all four human RD-only proteins are more similar to each other than to any of the yeast subunits, while in yeast Ego3p is not related to Ycr075wc-ap or Ynr034w-ap (). This suggests that early in evolution, ancestral Ragulator may have had one RD, which has duplicated repeatedly in fungi and metazoa, but it is also possible that there were two ancestral RDs, with gene conversion in the metazoal line. Neither Ycr075w-ap nor Ynr034w-ap has been studied directly, but our predictions may guide experiments into Ragulator function in a genetically tractable organism.
TRAPP: Prediction of a new LD in TRAPPC10 that may explain the Rab11-GEF activity of TRAPP-II
After finding that NPRL2 has an outlier LD distantly related to previously known LDs, we used that sequence to search for other LDs. The top hits matching the NPRL2 LD with probability of the same structure (pss) between 55% and 100% were (with one exception) either NPRL2 homologs (pss ≥ 99%, n = 10) or known members of the DENN superfamily (97 ≥ pss ≥ 57%, n = 16). The single exception was the N-terminus of human TRAPP-II subunit TRAPPC10. Residues 1–157 of TRAPPC10 aligned with NPRL2 (residues 1–142) with pss = 78%. Sequence conservation was maximal in the structural elements, and the largest difference was the insertion of a small extra β-sheet in TRAPPC10 between α2 and α3 (). The match to the NPRL2 LD was found in vertebrate TRAPPC10, but not in homologs from D. melanogaster, C. elegans, S. cerevisiae or plants. Forced alignment here showed that in these species TRAPPC10 lacks the β1, which weakens the overall alignments (Fig. S2A). We also checked if the LD is a general characteristic of TRAPP subunits, and although the N-termini of TRAPPC8 and TRAPPC9 have mixed α and β elements (Fig. S2B), the TRAPPC8 and TRAPPC9 sequences are so dissimilar from TRAPPC10 that a forced alignment could not be obtained between them and the NPRL2 LD. Therefore we predict a new LD only for TRAPPC10 in vertebrates, although for other species the possibility remains that β1 may be supplied from elsewhere in TRAPPC10.
Figure 4. A new LD at the N-terminus of human TRAPPC10 may dimerize with TRAPPC2. (A) Alignment of the N-termini of human NPRL2 (residues 1–142) and TRAPPC10 (residues 1–157). PSI-BLAST profiles for each sequence were aligned by HHalign.Citation27 Conserved and structural residues are colored according to the Clustalx scheme, gaps are indicated by dots, and predicted structural elements are boxed: helices red, sheets blue, with an extra sheet (β’) in TRAPPC10. (B) Diagram of the TRAPP-I heptamer,Citation12,Citation45 with the addition of the proposed LD dimerization between the N-terminus of TRAPPC10 (red) and TRAPPC2 (cyan). Other TRAPP-I subunits are: TRAPPC5 (yellow), TRAPPC3B (coral), TRAPPC4 (magenta), TRAPPC1 (green), TRAPPC3A (light pink), TRAPPC6 (blue). Rab1 (gray) binds TRAPPC4/C1 at a 45° angle from behind. The C-terminal extension of TRAPPC3A that produces GEF activity is indicated, and the PDZ domain in yeast TRAPPC4 is omitted. Subunit names are abbreviated to T-Cn. (C) Position of the TRAPPC2-TRAPPC10 dimer within TRAPP-II. The crystal structures of TRAPP-I (PDB: 3CUE and 2J3W)Citation12,Citation45 are superimposed on the TRAPP-II complex that was visualized by EM particle analysis.Citation49 Colors indicate the gross composition of TRAPP-II (for clarity only in the bottom half of the rotationally symmetrical particle): TRAPPC10 = magenta, TRAPPC9 = purple, C5orf44 = green, TRAPP-I core heptamer = red. Also indicated: a key residue in TRAPPC2 (Asp47, spheres).Citation45,Citation49 The N-terminus of TRAPPC10 is a model of NPRL2 (residues 1–142), which was created using Modeler using four known LD/RD structures.
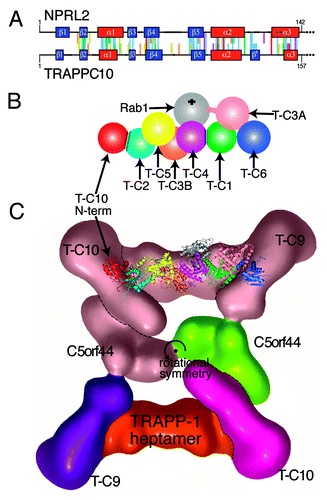
The discovery of any new domain in TRAPPC10 is significant because of TRAPP’s major role as a regulator of membrane traffic, but a new LD might specifically indicate an interaction with a GTPase. TRAPP is a highly conserved Rab1-GEF with several complexes (TRAPP-I/II/III) depending on the presence of optional subunits.Citation41 TRAPP-I is required for ER to Golgi traffic, tethers COP-II vesicles, and contains the six invariant short subunits that form the heptameric core for all TRAPP complexes (). Recently two extra-long subunits have been found in human TRAPP-I: TRAPPC11 and TRAPPC12 are widespread in eukaryotes, including fungi, but absent from budding yeast.Citation42 TRAPP-II is involved in post-Golgi traffic and contains the heptameric core and three optional long subunits: TRAPPC9, TRAPPC10 and C5orf44 (Trs120p, Trs130p and Trs65p in yeast).Citation43,Citation44 TRAPP-III is involved in autophagy, and contains a different optional long subunit TRAPPC8 (Trs85p in yeast). All three TRAPP complexes act as a GEF for Rab1 (Ypt1p), with the TRAPPC1/TRAPPC4 LD dimer forming a platform (), very similar to mglB ().Citation12 GEF activity derives from the extended C-terminus of one of the TRAPPC3 (Bet3p) subunits which invades the LD-LD-GTPase interface ().Citation12,Citation45
TRAPPC2 (also called sedlin, Trs20p in yeast) is a third TRAPP-I core subunit that contains an LD. Two structures of TRAPPC2 have been solved; one is a monomer, the other is a homodimer. The dimerization is facilitated in part by a conformational switch in TRAPPC2 ().Citation46 In TRAPP-I in the early secretory pathway, TRAPPC2 may homodimerize, and this form has been proposed to bind Sar1.Citation47 In TRAPP-II, the LD in TRAPPC10 provides a potential alternative partner LD for TRAPPC2 (). Although TRAPPC2 has been reported to interact with mammalian TRAPPC9,Citation48 this is inconsistent with a structural analysis of yeast TRAPP-II using EM of single particles, which placed the dimerization interface of TRAPPC2 adjacent to TRAPPC10 ().Citation49 We propose that the LD in TRAPPC10 dimerizes with TRAPPC2, which would provide an additional platform for GTPases in TRAPP-II compared with TRAPP-I (). This is interesting because it has been suggested that TRAPP-II in yeast is the GEF for Ypt31p or Ypt32p, homologs of human Rab11,Citation50,Citation51 although alternative explanations have been proposed.Citation52,Citation53 Since it is clear that Rab11 is not activated by the core TRAPP subunits,Citation50,Citation52 and there is no other good candidate for a specific Rab11-GEF (see DENNs, above), we predict that TRAPPC2 and TRAPPC10 form a second LD dimer in TRAPP-II that could interact with Rab11, and this prediction can be tested experimentally.
Conclusion
LDs and RDs are highly conserved interactors with small GTPases, mainly but not exclusively Rabs. While isolated single LD/RDs can interact with GTPases, usually a second domain is present, and most often this is another LD/RD. The characteristic side-by-side dimerization produces LD/RD platforms for regulators of membrane trafficking throughout evolution, with two major surfaces: the bottom, with four helices, which is often hydrophobic and may interact with membranes;Citation9 and the top, with two helices, that interacts with specific partners involved in membrane traffic – either GTPases or SNAREs.Citation5 The overall fold of LD/RDs is preserved with very little obvious sequence conservation,Citation11 so it is necessary to use remote homology searches that include secondary structure predictions to find new domains.Citation25 Such searches identified two RD-only proteins in yeast that are homologous to Ragulator subunits. We also identified an LD at the N-terminus of TRAPPC10 that may be part of a GTPase binding site unique to TRAPP-II.
| Abbreviations: | ||
| DENN | = | protein differentially expressed in normal cells and neoplasia |
| FLCN | = | folliculin |
| FNIP | = | folliculin interacting protein |
| GAP | = | GTPase activating protein |
| GEF | = | GDP:GTP exchange factor |
| HTH | = | helix turn helix |
| LD | = | longin domain |
| NPRL | = | nitrogen permease-like |
| pss | = | probability of the same structure |
| PDB | = | Protein Data Bank |
| RD | = | roadblock domain |
| SR | = | signal-recognition-particle receptor |
| TOR | = | target of rapamycin |
| TRAPP | = | transport protein particle |
Additional material
Download Zip (145.3 KB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Edina Harsay, Chris Loewen, Adam Grieve and Matt Hayes for helpful discussions. Our apologies to those whose work we could not cite through space considerations. Funding support was: R.D.D, Lowe Syndrome Trust; A.T.G, Marie Curie Initial Training Network “Sphingonet”; L.H.W., Medical Research Council (UK); F.A.B. and A.G., Wellcome Trust (Senior Investigator Award 097769/Z/11/Z).
Supplemental Material
Supplemental material may be found here: http://www.landesbioscience.com/journals/smallgtpases/article/24262/
References
- Yoshimura S, Gerondopoulos A, Linford A, Rigden DJ, Barr FA. Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J Cell Biol 2010; 191:367 - 81; http://dx.doi.org/10.1083/jcb.201008051; PMID: 20937701
- Gerondopoulos A, Langemeyer L, Liang JR, Linford A, Barr FA. BLOC-3 mutated in Hermansky-Pudlak syndrome is a Rab32/38 guanine nucleotide exchange factor. Curr Biol 2012; 22:2135 - 9; http://dx.doi.org/10.1016/j.cub.2012.09.020; PMID: 23084991
- Klöpper TH, Kienle N, Fasshauer D, Munro S. Untangling the evolution of Rab G proteins: implications of a comprehensive genomic analysis. BMC Biol 2012; 10:71; http://dx.doi.org/10.1186/1741-7007-10-71; PMID: 22873208
- Martinez-Arca S, Rudge R, Vacca M, Raposo G, Camonis J, Proux-Gillardeaux V, et al. A dual mechanism controlling the localization and function of exocytic v-SNAREs. Proc Natl Acad Sci U S A 2003; 100:9011 - 6; http://dx.doi.org/10.1073/pnas.1431910100; PMID: 12853575
- Schäfer IB, Hesketh GG, Bright NA, Gray SR, Pryor PR, Evans PR, et al. The binding of Varp to VAMP7 traps VAMP7 in a closed, fusogenically inactive conformation. Nat Struct Mol Biol 2012; 19:1300 - 9; http://dx.doi.org/10.1038/nsmb.2414; PMID: 23104059
- Sun Z, Anderl F, Fröhlich K, Zhao L, Hanke S, Brügger B, et al. Multiple and stepwise interactions between coatomer and ADP-ribosylation factor-1 (Arf1)-GTP. Traffic 2007; 8:582 - 93; http://dx.doi.org/10.1111/j.1600-0854.2007.00554.x; PMID: 17451557
- Wanschers B, van de Vorstenbosch R, Wijers M, Wieringa B, King SM, Fransen J. Rab6 family proteins interact with the dynein light chain protein DYNLRB1. Cell Motil Cytoskeleton 2008; 65:183 - 96; http://dx.doi.org/10.1002/cm.20254; PMID: 18044744
- Levivier E, Goud B, Souchet M, Calmels TP, Mornon JP, Callebaut I. uDENN, DENN, and dDENN: indissociable domains in Rab and MAP kinases signaling pathways. Biochem Biophys Res Commun 2001; 287:688 - 95; http://dx.doi.org/10.1006/bbrc.2001.5652; PMID: 11563850
- Kurzbauer R, Teis D, de Araujo ME, Maurer-Stroh S, Eisenhaber F, Bourenkov GP, et al. Crystal structure of the p14/MP1 scaffolding complex: how a twin couple attaches mitogen-activated protein kinase signaling to late endosomes. Proc Natl Acad Sci U S A 2004; 101:10984 - 9; http://dx.doi.org/10.1073/pnas.0403435101; PMID: 15263099
- Miertzschke M, Koerner C, Vetter IR, Keilberg D, Hot E, Leonardy S, et al. Structural analysis of the Ras-like G protein MglA and its cognate GAP MglB and implications for bacterial polarity. EMBO J 2011; 30:4185 - 97; http://dx.doi.org/10.1038/emboj.2011.291; PMID: 21847100
- Schlenker O, Hendricks A, Sinning I, Wild K. The structure of the mammalian signal recognition particle (SRP) receptor as prototype for the interaction of small GTPases with Longin domains. J Biol Chem 2006; 281:8898 - 906; http://dx.doi.org/10.1074/jbc.M512415200; PMID: 16439358
- Cai Y, Chin HF, Lazarova D, Menon S, Fu C, Cai H, et al. The structural basis for activation of the Rab Ypt1p by the TRAPP membrane-tethering complexes. Cell 2008; 133:1202 - 13; http://dx.doi.org/10.1016/j.cell.2008.04.049; PMID: 18585354
- Wu X, Bradley MJ, Cai Y, Kümmel D, De La Cruz EM, Barr FA, et al. Insights regarding guanine nucleotide exchange from the structure of a DENN-domain protein complexed with its Rab GTPase substrate. Proc Natl Acad Sci U S A 2011; 108:18672 - 7; http://dx.doi.org/10.1073/pnas.1110415108; PMID: 22065758
- Gong R, Li L, Liu Y, Wang P, Yang H, Wang L, et al. Crystal structure of the Gtr1p-Gtr2p complex reveals new insights into the amino acid-induced TORC1 activation. Genes Dev 2011; 25:1668 - 73; http://dx.doi.org/10.1101/gad.16968011; PMID: 21816923
- Jeong JH, Lee KH, Kim YM, Kim DH, Oh BH, Kim YG. Crystal structure of the Gtr1p(GTP)-Gtr2p(GDP) protein complex reveals large structural rearrangements triggered by GTP-to-GDP conversion. J Biol Chem 2012; 287:29648 - 53; http://dx.doi.org/10.1074/jbc.C112.384420; PMID: 22807443
- Zhang T, Péli-Gulli MP, Yang H, De Virgilio C, Ding J. Ego3 functions as a homodimer to mediate the interaction between Gtr1-Gtr2 and Ego1 in the ego complex to activate TORC1. Structure 2012; 20:2151 - 60; http://dx.doi.org/10.1016/j.str.2012.09.019; PMID: 23123112
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008; 320:1496 - 501; http://dx.doi.org/10.1126/science.1157535; PMID: 18497260
- Marat AL, Dokainish H, McPherson PS. DENN domain proteins: regulators of Rab GTPases. J Biol Chem 2011; 286:13791 - 800; http://dx.doi.org/10.1074/jbc.R110.217067; PMID: 21330364
- Nookala RK, Langemeyer L, Pacitto A, Ochoa-Montaño B, Donaldson JC, Blaszczyk BK, et al. Crystal structure of folliculin reveals a hidDENN function in genetically inherited renal cancer. Open Biol 2012; 2:120071; http://dx.doi.org/10.1098/rsob.120071; PMID: 22977732
- Baba M, Furihata M, Hong SB, Tessarollo L, Haines DC, Southon E, et al. Kidney-targeted Birt-Hogg-Dube gene inactivation in a mouse model: Erk1/2 and Akt-mTOR activation, cell hyperproliferation, and polycystic kidneys. J Natl Cancer Inst 2008; 100:140 - 54; http://dx.doi.org/10.1093/jnci/djm288; PMID: 18182616
- Li L, Kim E, Yuan H, Inoki K, Goraksha-Hicks P, Schiesher RL, et al. Regulation of mTORC1 by the Rab and Arf GTPases. J Biol Chem 2010; 285:19705 - 9; http://dx.doi.org/10.1074/jbc.C110.102483; PMID: 20457610
- Tsai PC, Lee SW, Liu YW, Chu CW, Chen KY, Ho JC, et al. Afi1p functions as an Arf3p polarization-specific docking factor for development of polarity. J Biol Chem 2008; 283:16915 - 27; http://dx.doi.org/10.1074/jbc.M802550200; PMID: 18397879
- Denef N, Chen Y, Weeks SD, Barcelo G, Schüpbach T. Crag regulates epithelial architecture and polarized deposition of basement membrane proteins in Drosophila. Dev Cell 2008; 14:354 - 64; http://dx.doi.org/10.1016/j.devcel.2007.12.012; PMID: 18331716
- Xiong B, Bayat V, Jaiswal M, Zhang K, Sandoval H, Charng WL, et al. Crag is a GEF for Rab11 required for rhodopsin trafficking and maintenance of adult photoreceptor cells. PLoS Biol 2012; 10:e1001438; http://dx.doi.org/10.1371/journal.pbio.1001438; PMID: 23226104
- Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 2005; 33:Web Server issue W244-8; http://dx.doi.org/10.1093/nar/gki408; PMID: 15980461
- Zhang D, Iyer LM, He F, Aravind L. Discovery of Novel DENN Proteins: Implications for the Evolution of Eukaryotic Intracellular Membrane Structures and Human Disease. Front Genet 2012; 3:283; http://dx.doi.org/10.3389/fgene.2012.00283; PMID: 23248642
- Levine TP, Daniels RD, Gatta AT, Wong LH, Hayes MJ. The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN Rab-GEFs. Bioinformatics 2013; 29:499 - 503; http://dx.doi.org/10.1093/bioinformatics/bts725; PMID: 23329412
- Morris HR, Waite AJ, Williams NM, Neal JW, Blake DJ. Recent advances in the genetics of the ALS-FTLD complex. Curr Neurol Neurosci Rep 2012; 12:243 - 50; http://dx.doi.org/10.1007/s11910-012-0268-5; PMID: 22477152
- Kinch LN, Grishin NV. Longin-like folds identified in CHiPS and DUF254 proteins: vesicle trafficking complexes conserved in eukaryotic evolution. Protein Sci 2006; 15:2669 - 74; http://dx.doi.org/10.1110/ps.062419006; PMID: 17075139
- Nordmann M, Cabrera M, Perz A, Bröcker C, Ostrowicz C, Engelbrecht-Vandré S, et al. The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr Biol 2010; 20:1654 - 9; http://dx.doi.org/10.1016/j.cub.2010.08.002; PMID: 20797862
- Carmona-Rivera C, Simeonov DR, Cardillo ND, Gahl WA, Cadilla CL. A divalent interaction between HPS1 and HPS4 is required for the formation of the biogenesis of lysosome-related organelle complex-3 (BLOC-3). Biochim Biophys Acta 2013; 1833:468 - 78; http://dx.doi.org/10.1016/j.bbamcr.2012.10.019; PMID: 23103514
- Kowalczyk MS, Hughes JR, Babbs C, Sanchez-Pulido L, Szumska D, Sharpe JA, et al. Nprl3 is required for normal development of the cardiovascular system. Mamm Genome 2012; 23:404 - 15; http://dx.doi.org/10.1007/s00335-012-9398-y; PMID: 22538705
- Neklesa TK, Davis RW. A genome-wide screen for regulators of TORC1 in response to amino acid starvation reveals a conserved Npr2/3 complex. PLoS Genet 2009; 5:e1000515; http://dx.doi.org/10.1371/journal.pgen.1000515; PMID: 19521502
- Dokudovskaya S, Waharte F, Schlessinger A, Pieper U, Devos DP, Cristea IM, et al. A conserved coatomer-related complex containing Sec13 and Seh1 dynamically associates with the vacuole in Saccharomyces cerevisiae. Mol Cell Proteomics 2011; 10:M110 - , 006478; http://dx.doi.org/10.1074/mcp.M110.006478; PMID: 21454883
- Wu X, Tu BP. Selective regulation of autophagy by the Iml1-Npr2-Npr3 complex in the absence of nitrogen starvation. Mol Biol Cell 2011; 22:4124 - 33; http://dx.doi.org/10.1091/mbc.E11-06-0525; PMID: 21900499
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 2010; 141:290 - 303; http://dx.doi.org/10.1016/j.cell.2010.02.024; PMID: 20381137
- Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell 2012; 150:1196 - 208; http://dx.doi.org/10.1016/j.cell.2012.07.032; PMID: 22980980
- Kogan K, Spear ED, Kaiser CA, Fass D. Structural conservation of components in the amino acid sensing branch of the TOR pathway in yeast and mammals. J Mol Biol 2010; 402:388 - 98; http://dx.doi.org/10.1016/j.jmb.2010.07.034; PMID: 20655927
- Wu B, Yee A, Ramelot T, Lemak A, Semesi A, Kennedy M, et al. Solution structure of hypothetical protein yst6499 from Saccharomyces cerevisiae. Northeast Structural Genomics Consortium Target YT727/ Ontario Center for Structural Proteomics Target yst6499; http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?uid=2GRG.
- Hall J, Song Y, Karplus PA, Barbar E. The crystal structure of dynein intermediate chain-light chain roadblock complex gives new insights into dynein assembly. J Biol Chem 2010; 285:22566 - 75; http://dx.doi.org/10.1074/jbc.M110.103861; PMID: 20472935
- Barrowman J, Bhandari D, Reinisch K, Ferro-Novick S. TRAPP complexes in membrane traffic: convergence through a common Rab. Nat Rev Mol Cell Biol 2010; 11:759 - 63; http://dx.doi.org/10.1038/nrm2999; PMID: 20966969
- Scrivens PJ, Noueihed B, Shahrzad N, Hul S, Brunet S, Sacher M. C4orf41 and TTC-15 are mammalian TRAPP components with a role at an early stage in ER-to-Golgi trafficking. Mol Biol Cell 2011; 22:2083 - 93; http://dx.doi.org/10.1091/mbc.E10-11-0873; PMID: 21525244
- Koumandou VL, Dacks JB, Coulson RM, Field MC. Control systems for membrane fusion in the ancestral eukaryote; evolution of tethering complexes and SM proteins. BMC Evol Biol 2007; 7:29; http://dx.doi.org/10.1186/1471-2148-7-29; PMID: 17319956
- Choi C, Davey M, Schluter C, Pandher P, Fang Y, Foster LJ, et al. Organization and assembly of the TRAPPII complex. Traffic 2011; 12:715 - 25; http://dx.doi.org/10.1111/j.1600-0854.2011.01181.x; PMID: 21453443
- Kim YG, Raunser S, Munger C, Wagner J, Song YL, Cygler M, et al. The architecture of the multisubunit TRAPP I complex suggests a model for vesicle tethering. Cell 2006; 127:817 - 30; http://dx.doi.org/10.1016/j.cell.2006.09.029; PMID: 17110339
- Jeyabalan J, Nesbit MA, Galvanovskis J, Callaghan R, Rorsman P, Thakker RV. SEDLIN forms homodimers: characterisation of SEDLIN mutations and their interactions with transcription factors MBP1, PITX1 and SF1. PLoS One 2010; 5:e10646; http://dx.doi.org/10.1371/journal.pone.0010646; PMID: 20498720
- Venditti R, Scanu T, Santoro M, Di Tullio G, Spaar A, Gaibisso R, et al. Sedlin controls the ER export of procollagen by regulating the Sar1 cycle. Science 2012; 337:1668 - 72; http://dx.doi.org/10.1126/science.1224947; PMID: 23019651
- Zong M, Wu XG, Chan CW, Choi MY, Chan HC, Tanner JA, et al. The adaptor function of TRAPPC2 in mammalian TRAPPs explains TRAPPC2-associated SEDT and TRAPPC9-associated congenital intellectual disability. PLoS One 2011; 6:e23350; http://dx.doi.org/10.1371/journal.pone.0023350; PMID: 21858081
- Yip CK, Berscheminski J, Walz T. Molecular architecture of the TRAPPII complex and implications for vesicle tethering. Nat Struct Mol Biol 2010; 17:1298 - 304; http://dx.doi.org/10.1038/nsmb.1914; PMID: 20972447
- Jones S, Newman C, Liu F, Segev N. The TRAPP complex is a nucleotide exchanger for Ypt1 and Ypt31/32. Mol Biol Cell 2000; 11:4403 - 11; PMID: 11102533
- Zou S, Liu Y, Zhang XQ, Chen Y, Ye M, Zhu X, et al. Modular TRAPP complexes regulate intracellular protein trafficking through multiple Ypt/Rab GTPases in Saccharomyces cerevisiae. Genetics 2012; 191:451 - 60; http://dx.doi.org/10.1534/genetics.112.139378; PMID: 22426882
- Wang W, Sacher M, Ferro-Novick S. TRAPP stimulates guanine nucleotide exchange on Ypt1p. J Cell Biol 2000; 151:289 - 96; http://dx.doi.org/10.1083/jcb.151.2.289; PMID: 11038176
- Chen S, Cai H, Park SK, Menon S, Jackson CL, Ferro-Novick S. Trs65p, a subunit of the Ypt1p GEF TRAPPII, interacts with the Arf1p exchange factor Gea2p to facilitate COPI-mediated vesicle traffic. Mol Biol Cell 2011; 22:3634 - 44; http://dx.doi.org/10.1091/mbc.E11-03-0197; PMID: 21813735
- Fan S, Wei Z, Xu H, Gong W. Crystal structure of human synbindin reveals two conformations of longin domain. Biochem Biophys Res Commun 2009; 378:338 - 43; http://dx.doi.org/10.1016/j.bbrc.2008.04.143; PMID: 18466758