Abstract
Live attenuated vaccines are adept in stimulating protective immunity. Methods for generating such vaccines have largely adopted strategies used with Salmonella enterica. Yet, when similar strategies were tested in other gram-negative bacteria, the virulence factors or genes responsible to incapacitate Salmonella often failed in providing the desired outcome. Consequently, conventional live vaccines rely on prior knowledge of the pathogen’s virulence factors to successfully attenuate them. This can be problematic since such bacterial pathogens normally harbor thousands of genes. To circumvent this problem, we found that overexpression of bacterial appendages, e.g., fimbriae, capsule, and flagella, could successfully attenuate wild-type (wt) Salmonella enterica serovar Typhimurium. Further analysis revealed these attenuated Salmonella strains conferred protection against wt S. Typhimurium challenge as effectively as genetically defined Salmonella vaccines. We refer to this strategy as attenuating gene expression (AGE), a simple efficient approach in attenuating bacterial pathogens, greatly facilitating the construction of live vaccines.
Introduction
Construction of live bacterial vaccines is traditionally dependent on mutating known virulence genes to adequately inactivate them. Since there are thousands of genes in a bacterial genome, screening for a suitable virulence gene to mutate can be a tedious process, and there is always the chance it may be inadequate to sufficiently reduce a pathogen’s virulence. Depending on the pathogen, a second mutation may be required to be introduced to avoid wild-type (wt) reversions or simply, the virulence gene targeted for inactivation results in an inadequate phenotype. Since bacteria have multiple virulence genes, it is often unclear how the remaining virulence genes will behave, and thus, upon inactivation of a given virulence gene, may have variable outcomes because of the varied impairment. For example, the Salmonella enterica serovar Typhimurium asd gene encodes aspartate-semialdehyde dehydrogenase, responsible for the formation of diaminopimelic acid (DAP), an essential component for gram-negative bacterial cell walls.Citation1 ∆asd S. Typhimurium cannot survive in regular medium, such as lysogeny broth (LB), unless LB is supplemented with DAP to facilitate cell wall formation. Since exogenous DAP is not normally found in the mammalian host, the ∆asd S. Typhimurium are unable to replicate within the host, and thus quickly become eliminated. Consequently, such strain was found to elicit a weak protective immune response in mice despite being given two consecutive oral doses with high inoculum.Citation2 Further development of other auxotrophic mutants, particularly the aro-based mutants with deficiencies in aromatic amino acid pathways, first in S. Typhi,Citation3 then adapted for S. Typhimurium with the aroA mutation, has led to successfully generated vaccines. The attenuated Salmonella strains by Hoiseth and StockerCitation4 has led to the extensive development of vaccines for typhoid fever in humansCitation5,Citation6 and salmonellosis in calves,Citation7-Citation9 pigs,Citation10-Citation13 and chickens.Citation14-Citation16 Moreover, such live vaccines also allow for the expression of heterologous genes from other pathogenic organisms that can stimulate both humoral and cell-mediated arms of immunity.Citation17-Citation19 When these are attenuated, one unique feature of live vector vaccines is their ability to stimulate both the mucosal and systemic immune compartments of the host while having limited capacity for growth and pathogenicity. Such attenuated mutants then allow for stimulation of pathogen recognition receptor pathways often required for protective immunity. When paired with the appropriate vaccine vector, heterologous passenger antigens (Ags) are also driven down similar innate and adaptive immune pathways resulting in similar host responses conferring protection against the heterologous pathogen. To be optimally protective, protection to the heterologous pathogen should share similar pathways as with those used by the vaccine vector for protection. In addition, such live vaccines can sustain Ag (vaccine) exposure by this self-generating delivery system. Having a live vaccine bearing two genetically dipartite mutations restricts the chance of in vivo reversion to a wt phenotype or regaining its virulence.Citation20,Citation21 Thus, a safe vaccine can then readily be developed.
Conventional methods of generating live attenuated vaccines are often dependent on the inactivation of virulence genes in one particular bacterial species, but its application does not always guarantee success in different species. Take ΔaroA S. Typhimurium as an example, this strain is a highly immunogenic and efficacious live vaccine, where one dose is generally sufficient to protect against wt S. Typhimurium challenge.Citation22 Likewise, another well-characterized live vaccine, ΔphoP S. Typhimurium,Citation23,Citation24 has been licensed for use in the poultry industry, and its corresponding serovar Typhi (Ty800) also appears safe and immunogenic in humans.Citation25,Citation26 However, the same inactivation strategy that proved successful in Salmonella failed to perform as effectively when adapted to other pathogens such as Yersinia and Brucella. Compared with ΔaroA S. Typhimurium, ΔaroA Yersinia enterocolitica 0:8 was less effective, requiring several doses to achieve measurable protection,Citation27 while the ΔphoP mutation in Y. pestis could only achieve a reduction in lethal dose by approximately 75-foldCitation28 vs. a fully attenuated ΔphoP S. Typhimurium. Although the ΔaroC Brucella suis was found to be attenuated,Citation29 when similarly applied to B. abortus, our laboratory found that ΔaroC B. abortus only conferred partial protection against wt Brucella challenge (unpublished data). These sharply contrasting results involving the same gene mutation in different bacterial species imply that genetic inactivation strategies are not always predictable on how they will perform in other bacterial species.
Conventional methods for developing live vaccines rely on the mutagenesis/deletion of one or more virulence genes. However, it is not always clear which virulence genes to target. Such mutagenesis approaches often have a negligible impact or conversely, will result in an overattenuated phenotype. In either case, the mutation will not have the desired outcome in modifying the bacterial pathogen resulting in its ineffectiveness as a possible vaccine. Thus, the selection of a suitable gene deemed appropriate for mutagenesis is often dependent on trial and error not knowing how much of the virulence will be sufficiently attenuated to be conducive as a live vaccine. Alternatively, if the virulence gene encodes a regulator, such as S. Typhimurium PhoP,Citation30 its deletion will affect an array of other genes, which will indirectly affect the immunogenicity of the mutant. Given these variables including extent of attenuation (overattenuated vs. underattenuated), vaccine persistence, heterologous vaccine stabilization, and the absence of antibiotic selection, it is often a delicate balance to derive new live, attenuated vaccines for bacterial pathogens. There is no real formula to successfully predict how the loss of function for a virulence gene will affect the overall virulence of the pathogen. Certainly, a method that can readily predict a vaccine’s performance could expedite vaccines for testing, and importantly, enable learning of what are the correlates of protective immunity. Because live vaccines mimic many aspects of natural infections, new methods are needed to quickly test vaccines while simultaneously learning aspects of the pathogens’ virulence. In seeking alternative methods to generate live vaccine mutants, in this review article, we introduce two novel methods for attenuating bacterial pathogens, the “regulated delayed attenuation” and the “attenuating gene expression (AGE)”, with particular emphasis on the recently developed AGE technology.
Regulated Delayed Attenuation
During the past two decades, a novel method of “regulated delayed attenuation” was created to attenuate bacterial pathogens.Citation31,Citation32 The premise of this approach is to use a promoter which can delay turning off the gene expression, particularly shutting off virulence genes after vaccination. This method exploits the capacity of allowing an initial “wild-type” infection to enable vaccination, but upon encountering a particular environment within the host, e.g., engulfed by a phagocytic cell, this triggers the live vaccine to begin shutting off selected virulence genes. The advantage of this approach is that there is no prerequisite virulence gene deletion or mutation except within the gene’s promoter allowing the bacteria to reproduce for a few generations during the initial infection. Following infection, the bacteria lose the capacity to reproduce due to their constrained expression of the virulence genes regulated by the modified promoter. Using this attenuation method, the substituted or modified promoter has a pivotal role in governing the real-time control of virulence gene expression. Theoretically, any promoter that results in the loss or the minimization of gene expression in vivo is considered as a “regulated, delayed attenuation” system. The promoter that is most frequently used is PBAD, which is derived from the arabinose operon which encodes for three enzymes (AraA, AraB, and AraC) needed to convert arabinose into a suitable metabolic form. Expression of PBAD is induced by the presence of arabinose, which is not normally found in the host and is supplied in the culture medium. Consequently, after infection, since arabinose is unavailable in vivo in the host, expression of PBAD will turn off, resulting in the loss of virulence gene expression. As a result, the pathogen is engulfed by the host phagocytic cells enabling an adaptive immune response to evolve. Attenuation in this dynamic fashion ultimately allows the development of a robust immune response conferring protection.Citation33 However, this technology still requires prior knowledge of the virulence genes to target.
AGE and Its Attenuation Mechanisms
To limit the dependence on prior knowledge of virulence genes responsible for the bacteria’s pathogenicity, we created a new method, called AGE.Citation34 We have shown AGE to be highly effective in attenuating bacterial pathogens without having prior knowledge of the pathogen’s virulence factors. The principle of AGE is to attenuate bacterial pathogens through overexpression of bacterial surface appendages. Bacterial appendages such as flagella, pili, needles, and capsules are located on the cell surface, and their expression is normally tightly controlled by the bacteria. The precise control of these appendages is dependent on whether they are needed. For example, when bacteria are grown in semisolid agar, flagella are expressed to facilitate swimming.Citation35 Through tight controls for their gene expression, bacteria need to use their energy stores efficiently. However, aside from direct controls or environmental stimuli to activate gene expression of its appendages, we suspect there are other mechanisms governing this whole process, which disallows these genes to be expressed constitutively. To test this hypothesis, colonization factor antigen I (CFA/I) fimbriae from enterotoxigenic E. coli (ETEC)Citation36 were overexpressed in wt S. Typhimurium.Citation37 We found that a striking attenuation effect was engendered in wt salmonellae when this operon was placed under control of the hybrid promoter PtetA~PpagC~PphoP. The wt S. Typhimurium expressing cfa/I were unable to reproduce within macrophages and unable to colonize mouse tissues.Citation34 Furthermore, when mice orally infected with 1 × 109 CFUs (~20 000 × LD50) of recombinant Salmonella expressing cfa/I, all survived. This result suggests that overexpression of CFA/I fimbriae has a robust attenuating effect, not inferior to the conventional attenuation methods relying on virulence gene deletion. Because attenuation from AGE does not require prior knowledge of virulence factors, it greatly facilitates engineering novel vaccine candidates.
In-depth investigation revealed that the CFA/I-mediated attenuation may be due to the overexpression of channels. CFA/I fimbria secretion apparatus is composed of four components: the tip protein CfaE, the shaft protein CfaB, the chaperone protein CfaA, and the usher protein CfaC. The fimbriae CfaBE () mediate attachment of ETEC to the small intestine during the initial stage of infection to facilitate colonization of the small intestine and subsequently cause disease upon secretion of its toxins.Citation38 Neutralizing antibodies to CFA/I fimbriae protect against disease.Citation36 To understand the cause for CFA/I’s attenuating effect upon Salmonella, we overexpressed the CFA/I components individually or in various combinations, CfaA, CfaC, CfaAC, CfaACE, and CfaABC in wt Salmonella.Citation34 We found that the overexpression of CfaAC (or as CfaACE or as CfaABC) produced a similar attenuating effect as did the overexpression of the intact cfa/I operon. This suggests that the combined overexpression of CfaA and CfaC is responsible for the Salmonella’s attenuation. Since expression of usher gene cfaC alone did not attenuate wt Salmonella unless combined with the overexpression of chaperone gene cfaA, this implies that without CfaA, the CfaC cannot form a functional channel. Thus, it is the overexpression of functional channels that accounts for Salmonella’s attenuation by CfaAC.Citation34
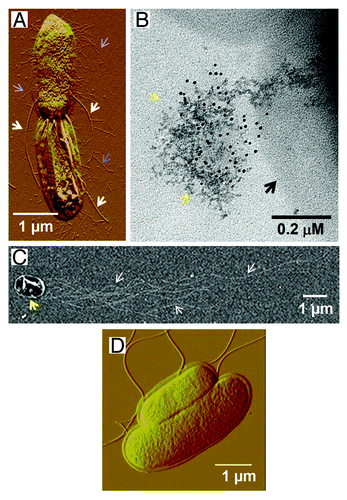
Figure 1. Images of AGE-modified Salmonella bearing different bacterial surface appendages. (A) Depicted is an image of S. Typhimurium P1-pHCCitation34 overexpressing ETEC CFA/I fimbriae as observed by atomic force microscopy. Narrow blue arrows indicate CFA/I fimbriae while bold white arrows indicate flagella. (B) Depicted is an image of S. Typhimurium P1-pHFCitation39 overexpressing Yersinia pestis F1 capsule as observed by field emission microscopy. The capsule is labeled with 20 nm immunogold particles (black spots). Narrow yellow arrows indicate the capsular proteins secreted by the cell forming a mushroom-like structure, which is in the early stage of capsule formation. Bold black arrows indicate the bacterial cell. (C) Depicted is an image of S. Typhimurium P1-pTP2DC+CCitation40 expressing flagella as observed by field emission microscopy. Narrow white arrows indicate flagella while bold yellow arrow indicates bacterial cell. (D) Depicted is an image of wt S. Typhimurium as observed by atomic force microscopy.
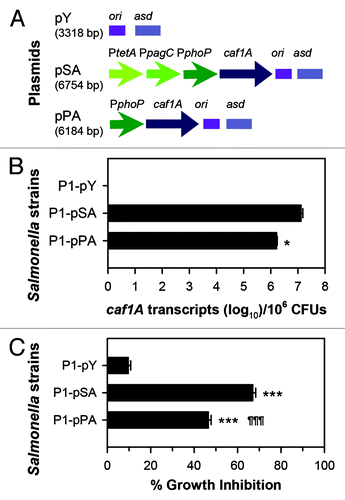
This conclusion is further supported by another finding that overexpression of the usher encoding gene caf1A alone is sufficient to attenuate wt Salmonella.Citation39 Yersinia pestis capsular secretion apparatus is composed of three components: the capsular protein Caf1, the usher protein Caf1A, and the chaperone-like protein Caf1M (). Overexpression of caf1A achieved a similar attenuation effect to that of overexpressing the whole caf operon in wt Salmonella.Citation39 Subsequent evaluation of overexpressed caf1A using a tripartite promoters (P1-pSA) or a single promoter in Salmonella (P1-pPA) was done (). Using real-time PCR, it was found that one-log greater number of transcripts for the Salmonella vaccine bearing the tripartite promoters () when compared with the Salmonella vaccine bearing a single PphoP. These differences in transcript levels correlated with increased sensitivity to growth inhibition upon treatment with erythromycin () suggesting that the Salmonella with the tripartite promoters have increased channel formation because of the enhanced sensitivity to the antibiotic treatment which in turn accounts for the Salmonella attenuation. Interestingly, we learned that Caf1A can self-assemble into functional channels in the outer membrane without the assistance of the chaperone Caf1M.Citation39 Further evidence supporting this hypothesis is evident from the erythromycin sensitivity assay whereby both CfaAC and Caf1A form functional channels in Salmonella since the osmotic permeability in these strains was significantly enhanced.Citation39
Figure 2. The extent of usher gene caf1A expression correlates to the amount of channel formation. (A) Schematic physical maps of asd-based plasmids are depicted: the caf1A gene is regulated by a tripartite fusion promoter in P1-pSACitation39 or a single promoter in P1-pPA; and the vector control lacking caf1A is harbored in P1-pY. (B) The expression of caf1A was determined by real-time polymerase chain reaction (RT-PCR). pSA DNA was used as a standard. The bacterial strains P1-pSA, -pPA, and control -pY were grown in LB medium at 37 °C with shaking at 200 rpm. Cells in log phase were harvested for RNA purification and verification of the number of transcripts using protocols according to the manufacturers' instructions (Invitrogen). The Student t-test was used to determine the difference of caf1A mRNA copy numbers between P1-pSA and -pPA, with *P < 0.05. (C) The Caf1A channel formation was determined by bacterial growth inhibition in the presence of erythromycin. The three strains P1-pSA, -pPA, and -pY were inoculated to LB medium at an optical density at 600 nm (OD600) of 0.2 with or without the supplementation of 64 µg/ml erythromycin. At three hours post-inoculation, cell OD600 was measured. Growth inhibition was calculated by using a formulation: 100 × (1 - OD600 with erythromycin/OD600 without erythromycin)%. The Tukey multiple comparison test was used to determine difference of growth inhibition among the three strains; ***P < 0.001 indicates the growth inhibition of P1-pSA or -pPA vs. -pY, and ¶¶¶P < 0.001 indicates the growth inhibition of -pPA vs. -pSA. Experiments for both (B) and (C) were performed three times. Presented data are the means ± SD.
In addition to the attenuation caused by the increased osmotic permeability, overexpression of the other proteins may also engender similar virulence inactivation. For instance, when the F1 capsular protein alone is overexpressed, the bacteria cannot secrete F1 to form a capsule on the cell surface due to the lack of channels. Thus, F1 was trapped and accumulated within the periplasmic space. Excessive F1 proteins in the periplasm may interfere with other periplasmic proteins impeding their function and possibly disrupting the integrity of the inner and outer membranes. Consequently, wt Salmonella was attenuated after overexpressing F1 capsular protein.Citation39 A similar attenuation effect was also achieved from the overexpression of Salmonella flagellin.Citation40 To regulate expression of flagellin constitutively, we used the chimeric promoter that was fused between the tetracycline resistance gene A promoter (PtetA) and the Salmonella phoP gene promoter (PphoP).Citation41 PtetA~PphoP was used to drive the co-expression of the flagella regulator encoding genes flhDC and the flagellin encoding gene fliC in wt Salmonella (). The viability of the flagellum overexpression Salmonella is dramatically reduced, and accordingly, its virulence is dampened.Citation40 Given these collective findings, overexpression of any gene products may not have an attenuating effect. This was demonstrated upon overexpression of either cfaA or cfaC alone which did not have any impact on wt Salmonella’s virulence.Citation34 In summary, our studies suggest that overexpression of appendage proteins is dependent on the formation of channels which represents an additional approach to attenuate wt gram-negative bacteria such as Salmonella.
The ultimate goal is to understand a pathogen’s virulence mechanisms, but this can be a tedious process particularly if the intent is to develop a defined mutant to become a sufficiently attenuated pathogen that ultimately can be used as a live vaccine. As evidence provided here, we show that overexpression of channel-dependent appendages provides an alternative method to attenuate a bacterial pathogen until the various virulence determinants can be elucidated. Results from our challenge studies using appendage-modified wt Salmonella revealed varying efficacy depending upon the degree of attenuation. For cfa/I operon using the attenuated Salmonella strains P1-pHC (wt S. Typhimurium strain P1 overexpressing the intact cfa/I operon under the control of PtetA~PpagC~PphoP), -pHcfaACE, -pHcfaABC, and -pHcfaAC exhibited 60%, 89%, 30%, and 60% protection, respectively, in BALB/c mice; and 33%, 80%, 50%, and 86% protection in C57BL/6 mice, respectively. Analysis of their virulence by both in vitro and in vivo assays revealed that P1-pHC and -pHcfaABC were more attenuated than -pHcfaACE and -pHcfaAC.Citation34 This became quite evident upon closer examination of splenic colonization in mice vaccinated with Salmonella strains P1-pHC and -pHcfaABC whereby these mice yielded no CFUs in their spleens unlike P1-pHcfaACE and -pHcfaAC vaccinated mice showed splenic colonization of 4880 and 1800 CFUs, respectively, at 5 d post-vaccination.Citation34 Thus, the P1-pHC and -pHcfaABC strains were overattenuated resulting in less protective efficacy unlike P1-pHcfaACE- and P1-pHcfaAC strains which were capable of eliciting a greater level of protection.Citation34 Similar differences in the extent of attenuation was also observed with our genetically defined mutants in Brucella.Citation42-Citation44
Strains were also developed that overexpressed flagella (). One strain that overexpressed the flagellar regulon master regulator genes, flhDC, and co-expressed flagellin gene, fliC is referred to as P1-pTP2DC+C, conferred 78% protection.Citation40 Without flhDC, the strain P1-pTP2fliC expressing fliC alone conferred only 33% protection.Citation40 P1-pTP2flhDC strain that expresses flhDC alone remained too virulent to be used as a live vaccine.Citation40 These results show that overexpression of fliC alone is sufficient to attenuate wt Salmonella unlike overexpression of flhDC. However, although co-expression of both fliC and flhDC in P1-pTP2DC+C exhibited similar attenuation in vivo as P1-pTP2fliC strain expressing fliC alone as measured by tissue colonization,Citation40 expression of flhDC is required for optimal immune protection since co-expression of fliC and flhDC is more protective than fliC alone. As with cfa/I and caf, the overexpressed FlhDC and FliC in P1-pTP2DC+C resulted in attenuation attributed to increased permeability of inner and outer membranes rendering this vaccine to become more immunogenic and allowing the recognition of protective antigens. With respect to the latter, flagellum is recognized by host TLR5, and because of this recognition, it is sometimes used as an adjuvant.Citation45,Citation46 Thus, its elevated expression may have enhanced the immunogenicity of the FlhDC-modified vaccine. In fact, the anti-flagella immune response elicited by P1-pTP2DC+C is significantly stronger than that induced by -pTP2fliC. Notably, some of these AGE-modified vaccines showed greater efficacy than the conventional aroA mutant vaccine, S. Typhimurium H647 (~70% protective), making these vaccine candidates suitable for further evaluation. Thus, the described AGE method is a useful approach in developing live bacterial vaccines.
Future Study and Potential Application of AGE
Since AGE is largely dependent on the overexpression of heterologous genes such as cfaAC and caf1A, one concern is these genes can undergo a reversion event reacquiring the wt genes when the host is infected with wt ETEC or Y. pestis, respectively, thus, compromising the vaccine’s attenuation capacity. Consequently, the bacterium will regain its virulence as a wt strain faltering the safety of this vaccine approach. To circumvent this limitation, we propose to introduce a hok/sok systemCitation47 to the bacteria in order to prevent the potential virulence reversion. The hok/sok system is a postsegregational killing mechanism employed by the R1 plasmid in E. coli. Once the R1 plasmid is lost, bacteria will be lysed by the toxic protein Hok.Citation48 Specifically, the hok/sok system would be placed under the control of an osmotic response promoter, such as promoter p1 of gene proP.Citation49 When cfaAC or caf1A is overexpressed in a bacterial organism, cell permeability will increase and thus p1 can turn on to synthesize an excessive amount of sok mRNA. As such, the killing mechanism by hok/sok will remain silent. However, once the cfaAC or caf1A is mutated, the bacterial osmotic permeability will recover to its normal status, and the osmotic promoter regulating sok gene expression will be switched off enabling hok gene expression. Once the killing function of the hok/sok system is activated, the cfaAC- or caf1A-mutated cells will then lyse. Thus, by introducing the hok/sok system into channel-overexpressed bacteria, the vaccine strain will not be able to regain virulence or assume a wt phenotype and will remain attenuated or be subsequently eliminated.
This AGE technology has a number of applications. First, because the level of Salmonella’s attenuation negatively correlates to their promoter potency,Citation34,Citation39 by selecting promoters with different strengths, we can then achieve varied levels of attenuation for the vaccine. This feature contrasts sharply with the traditional attenuating gene deletion/inactivation methods. Such gene inactivation methods usually generate vaccines whereby the mutant strain retains a “fixed” residual virulence. Thus, the safety of such a live vaccine depends upon how much residual virulence the bacteria possess to enable vaccination without compromising the host. Previous studies have shown that less attenuated strains generally confer greater protection, but generally these are less safe to use because of their remaining virulence. In contrast, while more attenuated strains are generally safe, these tend to be less effective in conferring protection. This is evident upon mutagenesis of a single virulence gene, znuA, and subsequently upon deletion of both virulence genes, znuA and purE, from wt Brucella abortus.Citation42-Citation44 Although the ∆znuA B. abortus mutant showed better protection than the double mutant, ∆znuA ∆purE B. abortus, the former is less safe because of the longer duration it remained within the host than the double mutant vaccine. Nevertheless, because AGE can adjust the degrees of attenuation by using different promoter strengths, the balance among attenuation, residual virulence, safety, and protection can be achieved. Current studies are in progress to develop a Salmonella candidate vaccine adequately expressing caf1A (unpublished results). Second, AGE can also be adapted for live vaccine construction using conventional deletion mutants rather than wt strains. Again, the selection of suitable promoters can have a potent attenuation effect, and can be used to enhance the vaccine’s safety. Finally, since gram-negative bacterial pathogens all share a similar membrane structure, the AGE approach is applicable to other bacterial pathogens. Such studies are currently being evaluated in our laboratory.
Conclusion
AGE is as effective as the conventional virulence gene deletion methods to inactivate bacterial pathogens. It is relatively simple to implement and has a number of flexible options. Because of these traits, AGE shows promise and has the potential for application in developing live vaccines, particularly for gram-negative bacteria. This process can accelerate the screening of various candidate vaccines, particularly for those pathogens whose virulence factors are not well-defined.
Acknowledgments
This work is supported by grants from National Institutes of Health Grant R21 AI-080960, R01 AI-41123, R01 AI-093372, and Office of Navy Research Award N00014-10-1-0946.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Galán JE, Nakayama K, Curtiss R 3rd. Cloning and characterization of the asd gene of Salmonella typhimurium: use in stable maintenance of recombinant plasmids in Salmonella vaccine strains. Gene 1990; 94:29 - 35; http://dx.doi.org/10.1016/0378-1119(90)90464-3; PMID: 2227450
- Piao HH, Tam VT, Na HS, Kim HJ, Ryu PY, Kim SY, et al. Immunological responses induced by asd and wzy/asd mutant strains of Salmonella enterica serovar Typhimurium in BALB/c mice. J Microbiol 2010; 48:486 - 95; http://dx.doi.org/10.1007/s12275-010-0023-z; PMID: 20799091
- Bacon GA, Burrows TW, Yates M. The effects of biochemical mutation on the virulence of Bacterium typhosum: the virulence of mutants. Br J Exp Pathol 1950; 31:714 - 24; PMID: 14801354
- Hoiseth SK, Stocker BA. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 1981; 291:238 - 9; http://dx.doi.org/10.1038/291238a0; PMID: 7015147
- Hone DM, Tacket CO, Harris AM, Kay B, Losonsky G, Levine MM. Evaluation in volunteers of a candidate live oral attenuated Salmonella typhi vector vaccine. J Clin Invest 1992; 90:412 - 20; http://dx.doi.org/10.1172/JCI115876; PMID: 1644914
- Tacket CO, Hone DM, Losonsky GA, Guers L, Edelman R, Levine MM. Clinical acceptability and immunogenicity of CVD 908 Salmonella typhi vaccine strain. Vaccine 1992; 10:443 - 6; http://dx.doi.org/10.1016/0264-410X(92)90392-W; PMID: 1609547
- Robertsson JA, Lindberg AA, Hoiseth S, Stocker BA. Salmonella typhimurium infection in calves: protection and survival of virulent challenge bacteria after immunization with live or inactivated vaccines. Infect Immun 1983; 41:742 - 50; PMID: 6347895
- Smith BP, Reina-Guerra M, Stocker BA, Hoiseth SK, Johnson E. Aromatic-dependent Salmonella dublin as a parenteral modified live vaccine for calves. Am J Vet Res 1984; 45:2231 - 5; PMID: 6395725
- Jones PW, Dougan G, Hayward C, Mackensie N, Collins P, Chatfield SN. Oral vaccination of calves against experimental salmonellosis using a double aro mutant of Salmonella typhimurium.. Vaccine 1991; 9:29 - 34; http://dx.doi.org/10.1016/0264-410X(91)90313-U; PMID: 2008797
- Alam J, Singh BR, Hansda D, Singh VP, Verma JC. Evaluation of aroA deletion mutant of Salmonella enterica subspecies enterica serovar Abortusequi for its vaccine candidate potential. Indian J Exp Biol 2009; 47:871 - 9; PMID: 20099460
- Trebichavsky I, Splichalova A, Rychlik I, Hojna H, Muneta Y, Mori Y, et al. Attenuated aroA Salmonella enterica serovar Typhimurium does not induce inflammatory response and early protection of gnotobiotic pigs against parental virulent LT2 strain. Vaccine 2006; 24:4285 - 9; http://dx.doi.org/10.1016/j.vaccine.2006.02.054; PMID: 16584815
- Barrow PA, Page K, Lovell MA. The virulence for gnotobiotic pigs of live attenuated vaccine strains of Salmonella enterica serovars Typhimurium and Enteritidis. Vaccine 2001; 19:3432 - 6; http://dx.doi.org/10.1016/S0264-410X(01)00065-2; PMID: 11348707
- Lumsden JS, Kennedy BW, Mallard BA, Wilkie BN. The influence of the swine major histocompatibility genes on antibody and cell-mediated immune responses to immunization with an aromatic-dependent mutant of Salmonella typhimurium.. Can J Vet Res 1993; 57:14 - 8; PMID: 8431799
- Van Immerseel F, De Buck J, De Smet I, Mast J, Haesebrouck F, Ducatelle R. The effect of vaccination with a Salmonella enteritidis aroA mutant on early cellular responses in caecal lamina propria of newly-hatched chickens. Vaccine 2002; 20:3034 - 41; http://dx.doi.org/10.1016/S0264-410X(02)00227-X; PMID: 12126917
- Methner U, Barrow PA, Martin G, Meyer H. Comparative study of the protective effect against Salmonella colonisation in newly hatched SPF chickens using live, attenuated Salmonella vaccine strains, wild-type Salmonella strains or a competitive exclusion product. Int J Food Microbiol 1997; 35:223 - 30; http://dx.doi.org/10.1016/S0168-1605(96)01236-6; PMID: 9105931
- Cooper GL, Venables LM, Woodward MJ, Hormaeche CE. Vaccination of chickens with strain CVL30, a genetically defined Salmonella enteritidis aroA live oral vaccine candidate. Infect Immun 1994; 62:4747 - 54; PMID: 7927750
- Saxena M, Coloe PJ, Smooker PM. Influence of promoter, gene copy number, and preexisting immunity on humoral and cellular responses to a vectored antigen delivered by a Salmonella enterica vaccine. Clin Vaccine Immunol 2009; 16:78 - 87; http://dx.doi.org/10.1128/CVI.00253-08; PMID: 19005022
- Xie C, He JS, Zhang M, Xue SL, Wu Q, Ding XD, et al. Oral respiratory syncytial virus (RSV) DNA vaccine expressing RSV F protein delivered by attenuated Salmonella typhimurium.. Hum Gene Ther 2007; 18:746 - 52; http://dx.doi.org/10.1089/hum.2007.053; PMID: 17696764
- Medina E, Guzmán CA, Staendner LH, Colombo MP, Paglia P. Salmonella vaccine carrier strains: effective delivery system to trigger anti-tumor immunity by oral route. Eur J Immunol 1999; 29:693 - 9; http://dx.doi.org/10.1002/(SICI)1521-4141(199902)29:02<693::AID-IMMU693>3.0.CO;2-V; PMID: 10064087
- Fouts TR, Tuskan RG, Chada S, Hone DM, Lewis GK. Construction and immunogenicity of Salmonella typhimurium vaccine vectors that express HIV-1 gp120. Vaccine 1995; 13:1697 - 705; http://dx.doi.org/10.1016/0264-410X(95)00106-B; PMID: 8719522
- Salomonsson E, Kuoppa K, Forslund AL, Zingmark C, Golovliov I, Sjöstedt A, et al. Reintroduction of two deleted virulence loci restores full virulence to the live vaccine strain of Francisella tularensis.. Infect Immun 2009; 77:3424 - 31; http://dx.doi.org/10.1128/IAI.00196-09; PMID: 19506014
- O’Callaghan D, Maskell D, Liew FY, Easmon CS, Dougan G. Characterization of aromatic- and purine-dependent Salmonella typhimurium: attention, persistence, and ability to induce protective immunity in BALB/c mice. Infect Immun 1988; 56:419 - 23; PMID: 3276625
- Galán JE, Curtiss R 3rd. Virulence and vaccine potential of phoP mutants of Salmonella typhimurium.. Microb Pathog 1989; 6:433 - 43; http://dx.doi.org/10.1016/0882-4010(89)90085-5; PMID: 2671582
- Hohmann EL, Oletta CA, Miller SI. Evaluation of a phoP/phoQ-deleted, aroA-deleted live oral Salmonella typhi vaccine strain in human volunteers. Vaccine 1996; 14:19 - 24; http://dx.doi.org/10.1016/0264-410X(95)00173-X; PMID: 8821644
- Garmory HS, Brown KA, Titball RW. Salmonella vaccines for use in humans: present and future perspectives. FEMS Microbiol Rev 2002; 26:339 - 53; PMID: 12413664
- Hohmann EL, Oletta CA, Killeen KP, Miller SI. phoP/phoQ-deleted Salmonella typhi (Ty800) is a safe and immunogenic single-dose typhoid fever vaccine in volunteers. J Infect Dis 1996; 173:1408 - 14; http://dx.doi.org/10.1093/infdis/173.6.1408; PMID: 8648213
- Bowe F, O’Gaora P, Maskell D, Cafferkey M, Dougan G. Virulence, persistence, and immunogenicity of Yersinia enterocolitica O:8 aroA mutants. Infect Immun 1989; 57:3234 - 6; PMID: 2777382
- Oyston PC, Dorrell N, Williams K, Li SR, Green M, Titball RW, et al. The response regulator PhoP is important for survival under conditions of macrophage-induced stress and virulence in Yersinia pestis.. Infect Immun 2000; 68:3419 - 25; http://dx.doi.org/10.1128/IAI.68.6.3419-3425.2000; PMID: 10816493
- Foulongne V, Walravens K, Bourg G, Boschiroli ML, Godfroid J, Ramuz M, et al. Aromatic compound-dependent Brucella suis is attenuated in both cultured cells and mouse models. Infect Immun 2001; 69:547 - 50; http://dx.doi.org/10.1128/IAI.69.1.547-550.2001; PMID: 11119550
- Kato A, Groisman EA, Howard Hughes Medical Institute. The PhoQ/PhoP regulatory network of Salmonella enterica.. Adv Exp Med Biol 2008; 631:7 - 21; http://dx.doi.org/10.1007/978-0-387-78885-2_2; PMID: 18792679
- Curtiss R 3rd, Wanda SY, Gunn BM, Zhang X, Tinge SA, Ananthnarayan V, et al. Salmonella enterica serovar typhimurium strains with regulated delayed attenuation in vivo.. Infect Immun 2009; 77:1071 - 82; http://dx.doi.org/10.1128/IAI.00693-08; PMID: 19103774
- Sun W, Roland KL, Kuang X, Branger CG, Curtiss R 3rd. Yersinia pestis with regulated delayed attenuation as a vaccine candidate to induce protective immunity against plague. Infect Immun 2010; 78:1304 - 13; http://dx.doi.org/10.1128/IAI.01122-09; PMID: 20086087
- Curtiss R 3rd, Xin W, Li Y, Kong W, Wanda SY, Gunn B, et al. New technologies in using recombinant attenuated Salmonella vaccine vectors. Crit Rev Immunol 2010; 30:255 - 70; http://dx.doi.org/10.1615/CritRevImmunol.v30.i3.30; PMID: 20370633
- Yang X, Suo Z, Thornburg T, Holderness K, Cao L, Lim T, et al. Expression of Escherichia coli virulence usher protein attenuates wild-type Salmonella.. Virulence 2012; 3:29 - 42; http://dx.doi.org/10.4161/viru.3.1.18447; PMID: 22286706
- Cao L, Suo Z, Lim T, Jun S, Deliorman M, Riccardi C, et al. Role of overexpressed CFA/I fimbriae in bacterial swimming. Phys Biol 2012; 9:036005; http://dx.doi.org/10.1088/1478-3975/9/3/036005; PMID: 22562964
- Yang X, Thornburg T, Holderness K, Suo Z, Cao L, Lim T, et al. Serum antibodies protect against intraperitoneal challenge with enterotoxigenic Escherichia coli. J Biomed Biotechnol 2011; 2011:632396; http://dx.doi.org/10.1155/2011/632396; PMID: 22007145
- Suo Z, Avci R, Deliorman M, Yang X, Pascual DW. Bacteria survive multiple puncturings of their cell walls. Langmuir 2009; 25:4588 - 94; http://dx.doi.org/10.1021/la8033319; PMID: 19260649
- Evans DG, Satterwhite TK, Evans DJ Jr., DuPont HL. Differences in serological responses and excretion patterns of volunteers challenged with enterotoxigenic Escherichia coli with and without the colonization factor antigen. Infect Immun 1978; 19:883 - 8; PMID: 346488
- Cao L, Lim T, Jun S, Thornburg T, Avci R, Yang X. Vulnerabilities in Yersinia pestis caf operon are unveiled by a Salmonella vector. PLoS One 2012; 7:e36283; http://dx.doi.org/10.1371/journal.pone.0036283; PMID: 22558420
- Yang X, Thornburg T, Suo Z, Jun S, Robison A, Li J, et al. Flagella overexpression attenuates Salmonella pathogenesis. PLoS One 2012; 7:e46828; http://dx.doi.org/10.1371/journal.pone.0046828; PMID: 23056473
- Yang X, Hinnebusch BJ, Trunkle T, Bosio CM, Suo Z, Tighe M, et al. Oral vaccination with salmonella simultaneously expressing Yersinia pestis F1 and V antigens protects against bubonic and pneumonic plague. J Immunol 2007; 178:1059 - 67; PMID: 17202369
- Yang X, Thornburg T, Walters N, Pascual DW. DeltaznuADeltapurE Brucella abortus 2308 mutant as a live vaccine candidate. Vaccine 2010; 28:1069 - 74; http://dx.doi.org/10.1016/j.vaccine.2009.10.113; PMID: 19914192
- Yang X, Skyberg JA, Cao L, Clapp B, Thornburg T, Pascual DW. Progress in Brucella vaccine development. Front Biol 2013; 8:60 - 77; http://dx.doi.org/10.1007/s11515-012-1196-0
- Yang X, Becker T, Walters N, Pascual DW. Deletion of znuA virulence factor attenuates Brucella abortus and confers protection against wild-type challenge. Infect Immun 2006; 74:3874 - 9; http://dx.doi.org/10.1128/IAI.01957-05; PMID: 16790759
- Mizel SB, Bates JT. Flagellin as an adjuvant: cellular mechanisms and potential. J Immunol 2010; 185:5677 - 82; http://dx.doi.org/10.4049/jimmunol.1002156; PMID: 21048152
- Mizel SB, Graff AH, Sriranganathan N, Ervin S, Lees CJ, Lively MO, et al. Flagellin-F1-V fusion protein is an effective plague vaccine in mice and two species of nonhuman primates. Clin Vaccine Immunol 2009; 16:21 - 8; http://dx.doi.org/10.1128/CVI.00333-08; PMID: 18987167
- Pecota DC, Kim CS, Wu K, Gerdes K, Wood TK. Combining the hok/sok, parDE, and pnd postsegregational killer loci to enhance plasmid stability. Appl Environ Microbiol 1997; 63:1917 - 24; PMID: 9143123
- Gerdes K, Thisted T, Martinussen J. Mechanism of post-segregational killing by the hok/sok system of plasmid R1: sok antisense RNA regulates formation of a hok mRNA species correlated with killing of plasmid-free cells. Mol Microbiol 1990; 4:1807 - 18; http://dx.doi.org/10.1111/j.1365-2958.1990.tb02029.x; PMID: 1707122
- Mellies J, Wise A, Villarejo M. Two different Escherichia coli proP promoters respond to osmotic and growth phase signals. J Bacteriol 1995; 177:144 - 51; PMID: 8002611