Abstract
Generation of a live attenuated vaccine for bacterial pathogens often requires prior knowledge of the pathogen’s virulence factors. We hypothesized an alternative approach of heterologous gene expression would make a wild-type (wt) pathogen more susceptible to host cell killing, thus, resulting in immunization. As proof of concept, the heterologous expression of enterotoxigenic E. coli (ETEC) colonization factor antigen I (CFA/I) was tested to attenuate Salmonella. The overexpression of CFA/I resulted in significant attenuation of wt Salmonella. In-depth studies revealed the attenuation depended on the co-expression of chaperone (CfaA) and usher (CfaC) proteins. Remarkably, the CfaAC-attenuated Salmonella conferred protection against wt Salmonella challenge. Mechanistic study indicated CfaAC made Salmonella outer membranes permeable, causing Salmonella to be vulnerable to host destruction. Thus, enhancing bacterial permeability via CfaAC represents an alternative method to attenuate pathogens despite the presence of unknown virulence factors.
Introduction
Salmonella enterica is an enteric bacterial pathogen responsible for a variety of food- and water-borne illnesses, ranging from gastroenteritis to typhoid fever,Citation1 and it colonizes a wide range of animal hosts, including humans.Citation2 Among more than 2,500 identified serovars,Citation3S. typhi alone is estimated to cause approximately 16 million cases annually, resulting in 600,000 deaths worldwide,Citation4 with 600 of the fatalities occurring in the USA.Citation5
S. typhimurium possesses a number of virulent determinants that facilitate its ability to colonize, invade host intestinal epithelia and spread systemically.Citation6 Conventional attenuation methods rely on mutating known or suspect virulent genes, and in turn, these attenuated S. typhimurium strains are then used as live vaccines. A well-known mutant strain is the ΔaroA S. typhimurium that can effect protection with a single dose against wt S. typhimurium challenge.Citation7 In the same vein, another mutant, ΔphoP S. typhimurium,Citation8 has been licensed for the poultry industry. However, the genetic inactivation strategies successfully used for Salmonella are not always adaptable to other pathogens, thus, requiring additional investigation to identify suspected virulence genes.
CFA/I fimbria is one of the most prevalent adhesins associated with human-specific ETEC fimbriae.Citation9 These fimbriae project from the bacterial surface to dock with specific receptors on the host cell surface within preferred nichesCitation10 to mediate small intestinal adherence and the elaboration of (heat stable and heat labile) enterotoxins that induce both fluid and electrolyte secretion. Four structural genes encode four proteins, which coordinate to form CFA/I fimbriae. cfaB encodes the ~15 kDa major subunit protein that forms the pili.Citation11 cfaE encodes the minor subunit protein, which locates to the tip of the fimbriae and is also responsible for nucleating fiber formation.Citation12 Adhesive phenotypes have been attributed to both the major and minor subunits of CFA/I fimbriae.Citation13 cfaA encodes the periplasmic chaperone that promotes subunit folding and transports the subunits to an outer membrane usher where ordered assembly of the filamentous heteropolymer is achieved.Citation12 cfaC encodes the usher believed to facilitate translocation of pilus subunits across the outer membrane.Citation14
In the current study, we propose an alternative strategy for attenuating Gram-negative bacteria via a process we refer to as attenuating gene expression (AGE), a method of heterologous gene overexpression tested with ETEC cfa/I operon. Inference to such a possibility has been suggested in previous work in which overexpression of cfa/I operon by an attenuated Salmonella strain alters host immune responses initially with a potent Th2 cell response to CFA/I fimbriaeCitation15,Citation16 and a lack of proinflammatory cytokine production by infected macrophages.Citation17 Such altered host responses have been determined to be beneficial against autoimmune diseases, whereby the Salmonella-CFA/I vaccine stimulates diverse regulatory T cell subsets.Citation18,Citation19 These unconventional findings suggest heterologous gene expression of cfa/I operon is a dynamic tool that can be alternatively explored either to modulate the host immune response or to attenuate the virulence of recombinant bacteria. To investigate the impact of cfa/I operon overexpression upon wt pathogens, S. typhimurium H71 strainCitation20 has been selected as a model bacterial pathogen to assess whether expression of ETEC cfa/I operon can attenuate wt Salmonella. The results show that indeed cfa/I operon expression attenuates wt Salmonella and is attributed primarily to the heterologous expression of the usher gene CfaC. This work demonstrates that it is feasible to attenuate Gram-negative bacteria using CfaC.
Results
Recombinant S. typhimurium H71 strains overexpressing CFA/I fimbriae
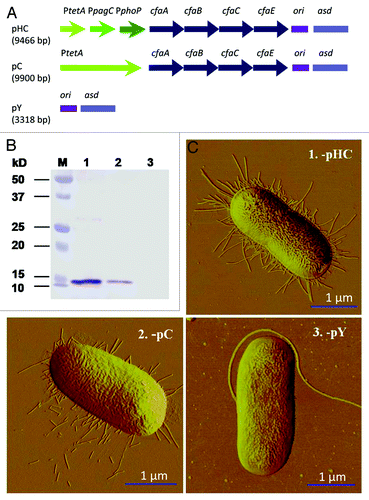
A benefit of utilizing S. typhimurium to assess the heterologous expression of fimbriae is the availability of reagents to enable stable heterologous gene expression using a well-established approach by introducing a mutation in its asd gene,Citation21 as previously shown for stable cfa/I operon expression.Citation22 This balanced-lethal mutation eliminates possible inactivation or loss of unstable variants that could compromise analysis of the described complemented strains, requiring retention of asd-based plasmids for S. typhimurium survival. Consequently, the strain Δasd::kanR S. typhimurium H71 (P1) was developed and transformed with plasmids pHC, pC and pY (), respectively, to obtain the recombinant strains P1-pHC, -pC and -pY. Expression of cfa/I operon was regulated by PtetACitation22 and PtetA~PpagC~PphoPCitation23 in pC and pHC, respectively, and empty vector pY served as a control. Protein gel blot and atomic force microscopy (AFM) analyses revealed the influence of the fused promoter on CFA/I fimbria expression. While P1-pY showed no detectable band, both -pHC and -pC showed CfaB expression (). Densitometric scanning revealed the amount of CfaB produced by P1-pHC was ~3.5-fold greater than that produced by the -pC strain, suggesting that fimbrial expression was enhanced by the fusion promoters when compared with the PtetA promoter alone. This was further confirmed by AFM that P1-pHC expressed more fimbriae than -pC strain; no fimbriae were observed for negative control -pY ().
Figure 1. Evaluation of CFA/I fimbriae expression by protein gel blot analysis and AFM imaging. (A) Schematic physical maps of asd-based plasmids. The cfa/I operon is regulated by a tripartite fusion promoter in P1-pHC, a single promoter in P1-pC and no cfa/I operon is harbored in control P1-pY. (B) Protein gel blot analysis shows CFA/I fimbria expression by P1-pHC (Lane 1) and -pC (Lane 2) and no fimbriae expression was detected for -pY (Lane 3). Prestained molecular weight standards are shown in Lane M. (C) AFM images indicate expression of CFA/I fimbriae for strains (1) P1-pHC and (2) -pC, but no fimbriae were observed for control strain (3) -pY.
P1-pHC and -pC are highly attenuated in vitro and in vivo
To investigate whether overexpression of CFA/I attenuates wt Salmonella, RAW264.7 macrophages were infected at varying ratios with P1-pHC, -pC and -pY. As depicted, P1-pHC was unable to survive in macrophages, regardless of the infection dose, and these salmonellae showed as much as 1000-fold reduction by 24 h (). Compared with P1-pY, -pC showed limited replication/survival to the original infection dose. These results show cfa/I operon expression impedes the salmonellae survival in macrophages.
Figure 2. Overexpression of cfa/I attenuates Salmonella and confers protective immunity. (A) Strains P1-pHC, -pC and -pY were assessed for their survival in RAW264.7 macrophages at varying bacteria to macrophage ratios of (1) 1:1, (2) 10:1 or (3) 100:1. While initial levels of infection (t = 0 h) were significantly different for each strain, at 4 or 24 h post-infection, survival rates were augmented for each tested strain. In fact, at 4 and 24 h post-infection, P1-pHC was cleared at or below the detection limit. While P1-pC was not cleared as efficiently, it still was impaired relative to -pY. Values are the mean ± SEM of three independent experiments. Differences in macrophage colonization were calculated using Tukey Kramer multiple comparisons test by P1-pY vs. (vs) -pHC or -pC are indicated as: **p < 0.01, ***p < 0.001. (B) Overexpression of cfa/I reduces tissue inflammation and colonization. BALB/c mice were orally dosed with 1 × 109 CFUs of P1-pHC, -pC, -pY, or wt strain H71 and at 5 d post-infection, (1) splenic weights and total CFUs/tissue were determined for (2) spleens, (3) PPs and (4) livers. Results are mean ± SEM of two independent experiments. Differences in tissue weight and colonization burden were calculated using Tukey Kramer multiple comparisons test. Values representing significant differences in weight or colonization in mice infected with wt H71 vs P1-pHC, -pC, or -pY are indicated as: *p < 0.05, **p < 0.01 and ***p < 0.001; ND, none detected. (C) cfa/I-attenuated Salmonella confers protection against wt Salmonella challenge. Groups of BALB/c mice were orally dosed with 1 × 109 CFUs of (1) P1-pHC (n = 15), -pC (n = 15), -pY (n = 15), wt H71 (n = 10), or 5 × 109 CFUs of H647 (n = 10); all mice given P1-pY, -pC, or wt H71 succumbed to infection. The Kaplan-Meier method was used to obtain the survival fractions for wt H71, P1-pHC, -pC and H647 administered mice. Survival fractions of wt H71 were compared with mice dosed with P1-pHC, -pC and H647 and significance was determined: **p < 0.01; and mice administered with P1-pY were compared with mice dosed with P1-pHC, -pC and H647 and significance was determined: ¶¶¶p < 0.001. Data are the mean from three experiments. (2) P1-pHC-immunized mice were challenged with 5 × 107 CFUs wt H71 strain and 60% of the mice survived with similar survival to H647-immunized mice at 70%. All sPBS-dosed mice succumbed to challenge. The Kaplan-Meier method was used to obtain the survival fractions after challenge for sPBS-, P1-pHC- and H647-immunized mice. Survival fractions of sPBS group were compared with P1-pHC and H647-immunized groups and significance was determined: ***p < 0.001. Data are the mean from two experiments. (D) A kinetic analysis of serum IgG and fecal IgA (1) anti-CFA/I fimbriae and (2) anti-heat-killed S. typhimurium (HKST) Ab titers were performed for BALB/c mice vaccinated with P1-pHC. Differences were calculated by Student t-test. Anti-CFA/I fimbriae IgG titers by P1-pHC immunized mice were significantly different from IgG titers by sPBS-dosed mice and indicated as: *p < 0.05, **p < 0.01 and ***p < 0.001; and anti-CFA/I fimbriae IgA titers by P1-pHC immunized mice vs sPBS-dosed mice are indicated as: ¶p < 0.05, ¶¶p < 0.01 and ¶¶¶p < 0.001. H647 induced no anti-CFA/I fimbriae titers, but it induced significantly higher anti-heat killed Salmonella typhimurium (HKST) IgG titers than sPBS-dosed mice and indicated as: †p < 0.05, ††p < 0.01 and †††p < 0.001; and IgA titers by H647 vs sPBS-dosed mice are indicated as: ‡p < 0.05, ‡‡p < 0.01 and ‡‡‡p < 0.001. Data are the mean ± SEM of two independent experiments.
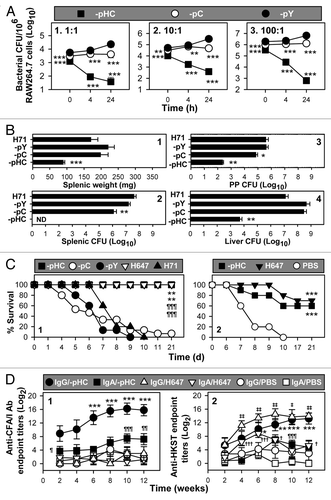
To test whether P1-pHC is attenuated in vivo, mouse colonization studies were performed. At 5 d after oral administration, spleens were weighed, and bacterial CFUs in the spleens, Peyer’s patches (PPs) and livers were enumerated (). The results showed no differences between P1-pY and wt strain H71 in splenic weights and tissue colonization, suggesting P1-pY is as virulent as H71. Splenic weights exhibited by P1-pHC infected mice were significantly reduced relative to -pC, -pY and H71 infected mice, but splenic weights among P1-pC, -pY and H71 infected mice were not significantly different from each other. Importantly, P1-pHC did not colonize the spleen () regardless of time point examined (Fig. S1), a phenomenon similar to phoP and rfaC Salmonella ser Enteritidis mutants,Citation24 suggesting -pHC was greatly attenuated. PP and liver CFUs of P1-pHC infected mice were also significantly less than -pC, -pY and wt H71 infected mice by 3,000–100,000-fold (). Splenic colonization by P1-pC infected mice was slightly reduced when compared with -pY or H71 infected mice, demonstrating less attenuation.
P1-pHC confers protection against wt S. typhimurium challenge
To assess the virulence of these three strains, groups of BALB/c mice were orally gavaged with P1-pHC, -pC, -pY, wt H71 and ΔaroA S. typhimurium vaccine strain H647.Citation20 Only those mice given P1-pHC or H647 survived, while only one of 15 mice in the -pC administered group survived and none of the -pY or H71 infected mice survived (). These results further demonstrate that enhanced expression of cfa/I operon attenuates Salmonella in vivo.
Since all P1-pHC and H647 immunized mice survived, these and a group of sterile phosphate buffered saline (sPBS)-dosed mice were orally challenged with wt H71 (). All sPBS-dosed mice succumbed to infection within 9 d, while 60% and 70% of the mice dosed with P1-pHC or H647 survived, respectively. The percentage of survival by the mice vaccinated with the H647 was similar to that previously described.Citation20,Citation25 Additional studies were conducted to assess the influence of the fusion promoter used in P1-pHC. The P1-pHV strain was constructed and found to be as virulent as P1-pY (Fig. S2). Thus, these results show P1-pHC is capable of conferring protective immunity against wt Salmonella challenge.
To determine if the expressed fimbriae were immunogenic in mice orally dosed with P1-pHC, H647 and sPBS, serum IgG and mucosal IgA anti-CFA/I fimbriae and anti-salmonellae endpoint antibody (Ab) titers were measured. Elevated IgG titers were induced to both CFA/I fimbriae and salmonellae when assessed 6 weeks after oral immunization with P1-pHC (). In addition, significant mucosal IgA anti-CFA/I fimbriae Abs were elicited shortly 2 weeks after oral immunization, and mucosal IgA anti-salmonellae Abs were elicited 8–10 weeks post-immunization. As expected, strain H647 induced no anti-CFA/I immunity, but stimulated elevated anti-salmonellae immunity. These results show P1-pHC is able to stimulate both systemic and mucosal Abs to CFA/I fimbriae as well to the live vaccine vector.
Expression of CfaC is essential for Salmonella attenuation
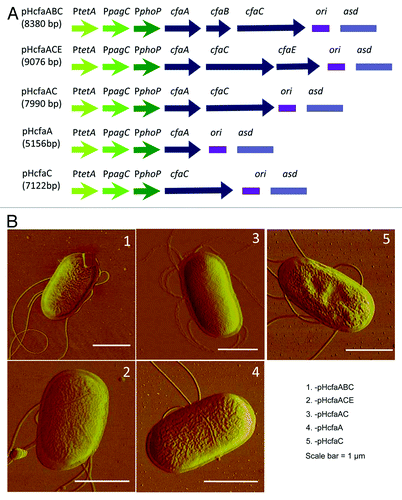
To determine which of the four genes in cfa/I operon was responsible for the observed attenuation, five cfa/I deletion mutants were generated (). cfaB or cfaE, or both, was deleted in-frame from pHC and referred to as pHcfaACE, pHcfaABC and pHcfaAC, respectively. The cfaBCE deletion mutant was pHcfaA, and the cfaABE deletion mutant was pHcfaC. No fimbriae were found by AFM on the bacterial cell surface (), indicating that both the major fimbrial subunit gene, cfaB, and the minor fimbria subunit gene, cfaE, are required for the formation of CFA/I fimbriae.
Figure 3. Schematic maps and AFM imaging of P1 strains bearing cfa/I gene components. (A) Schematic maps of five recombinant plasmids. (B) AFM images of P1 strains bearing the plasmid with (1) pHcfaABC, (2) pHcfaACE, (3) pHcfaAC, (4) pHcfaA or (5) pHcfaC. No fimbriae were detected in any of these strains.
P1-pHcfaABC, -pHcfaACE, -pHcfaAC, -pHcfaC, -pHcfaA and -pY were each evaluated for virulence in RAW264.7 cells at a bacteria-to-macrophage infection ratio of 1:1 (). P1-pHcfaA and -pHcfaC replicated within the macrophages as vigorously as -pY at 24 h post-infection. However, P1-pHcfaABC, -pHcfaACE and -pHcfaAC were unable to infect as robustly as -pY and, importantly, could not reproduce within macrophages. These results suggest either CfaB or CfaE may be dispensable for Salmonella attenuation in macrophages since their absence from -pHcfaAC resulted in attenuation. While the chaperone CfaA alone or the usher CfaC alone also could not induce Salmonella attenuation, the co-expression of CfaAC was essential since Salmonella attenuation was obtained for P1-pHcfaACE, -pHcfaABC or -pHcfaAC. In addition, co-expression of one of the structural proteins, CfaB or CfaE, may be important since colonization by P1-pHcfaABC and -pHcfaACE was 21.1- and 1.8-fold less than that of -pHcfaAC at 24 h, respectively.
Figure 4. The combination of cfaAC genes is required for Salmonella attenuation. (A) Evaluation of in vitro virulence in RAW264.7 macrophages was determined for P1 strains bearing the plasmids, -pHcfaABC, -pHcfaACE, -pHcfaAC, -pHcfaC, -pHcfaA or -pY. Macrophages were infected at a bacteria-to-macrophage ratio of 1:1 and the bacterial CFUs were enumerated at 0, 4 and 24 h post-infection. Differences in macrophage colonization were calculated using Tukey Kramer multiple comparisons test. Values depict the mean ± SEM of 4 independent experiments; ***p < 0.001 depicts significant differences in colonization vs P1-pY; ¶¶¶p < 0.001 depicts significant differences at 4 and 24 h post-infection for grouped strains, P1-pY, -pHcfaA and -pHcfaC vs P1-pHcfaAC, -pHcfaACE and -pHcfaABC. (B) Evaluation of in vivo virulence of cfa/I gene-expression of Salmonella strains was determined in BALB/c mice. Groups of BALB/c mice (5/group) were orally infected with one of the six strains (1 x 109 CFUs/strain/mouse) and at 5 d post-infection, (1) splenic weights and total CFUs/tissue from (2) spleens, (3) PPs and (4) livers were determined. Differences in tissue weight and colonization burden were calculated using Tukey Kramer multiple comparisons test. *p < 0.05, **p < 0.01 and ***p < 0.001 depict significant differences in splenic weights or splenic, PP and liver CFUs from mice infected with cfa/I gene-attenuated strains vs P1-pY infected mice. (C) To assess the lethality of the recombinant Salmonella strains, groups of mice were orally dosed with one of the 6 strains, as described above, in (B). Strains P1-pY (n = 10), -pHcfaA (n = 10) and -pHcfaC (n = 10) were lethal to BALB/c mice; the P1-pHcfaABC (n = 15) and -pHcfaAC (n = 15) infected mice survived and 90% of the P1-pHcfaACE (n = 10) infected mice survived. The Kaplan-Meier method was used to obtain the survival fractions. Values are the mean ± SEM of two (P1-pHcfaACE, -pHcfaA, -pHcfaC and -pY) or three (P1-pHcfaABC and -pHcfaAC) independent experiments; survival fractions obtained from mice dosed with P1-pHcfaABC, -pHcfaACE, or -pHcfaAC were compared with mice dosed with P1-pY, -pHcfaA, -pHcfaC and significance is shown: ***p < 0.001. Arrows indicate the time of infection.
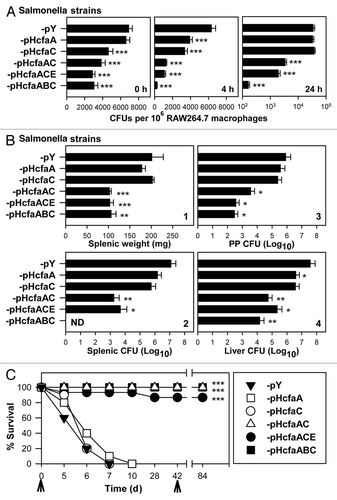
To assess the influence of CfaAC upon virulence in vivo, mice were orally infected with 1 × 109 CFUs of P1-pHcfaABC, -pHcfaACE, -pHcfaAC, -pHcfaC, -pHcfaA and -pY, and tissue and colonization were analyzed at 5 d post-infection (). As with the infection of the RAW264.7 macrophages, a segregation appeared among the recombinant strains: mice infected with P1-pY, -pHcfaA and -pHcfaC showed enlarged spleens and elevated CFUs in spleens, PPs and livers, while the mice infected with P1-pHcfaABC, -pHcfaACE and -pHcfaAC showed minimal inflammation of their spleens and low levels of salmonellae. Interestingly, no CFUs could be recovered from spleens of mice infected with P1-pHcfaABC, but salmonellae were recovered from PPs and livers. P1-pHcfaACE or -pHcfaAC appeared attenuated since between 2,500- and 6,900-fold less salmonellae than P1-pY could be recovered from their spleens. These results show P1-pHcfaABC, -pHcfaACE and -pHcfaAC are significantly attenuated by the co-expression of CfaAC, either in the presence or absence of the fimbrial subunits, CfaB or CfaE, and the expression of CfaC or CfaA alone is insufficient, and their virulence resembles control P1-pY.
Subsequent to these studies, the six strains were further evaluated for their extended virulence in BALB/c mice. Groups of mice were orally infected with one of the six strains as previously described. All mice infected with P1-pHcfaABC or -pHcfaAC survived, while 90% of -pHcfaACE infected mice survived (). In contrast, all mice infected with P1-pHcfaA, -pHcfaC or -pY succumbed to infection within 10 d. A second dose of P1-pHcfaABC, -pHACE or -pHcfaAC was given to the respective groups on day 42. No additional attrition to vaccination was observed. These results further confirm that, although CfaA or CfaC alone is unable to attenuate Salmonella, the co-expression of both or in combination with a fimbrial subunit attenuates wt Salmonella, even when given at a relatively high dose.
Analysis of the potential mechanisms involved in the Salmonella attenuation
The above studies illustrated that overexpression of either CFA/I fimbriae or the synergistic overexpression of CfaAC was able to inactivate the bacterial host in vitro and in vivo. We questioned whether the attenuation was due to the metabolic burden incurred from the expression of heterologous genes, as previously noticed.Citation26 Thus, we compared the growth rates of the seven strains, P1-pHcfaA, -pHcfaC, -pHcfaAC, -pHcfaABC, -pHcfaACE, -pHC and -pY. P1-pHC exhibited significantly slower growth than that of -pY, as early as 0.5 h post-inoculation, and it maintained the slowest growth rate to stationary phase among all the tested strains (Fig. S3). Since P1-pHC expressed all cfa/I components, its slow growth implies that the metabolic burden from overexpression of cfa/I operon impacts negatively upon the salmonellae, which may include the attenuation effects to the bacterial virulence. Growth rate of P1-pHcfaABC also showed a similar trend to -pHC, while -pHcfaA and -pHcfaC yielded slow growth rates at the stationary phase. However, the P1-pHcfaAC and -pHcfaACE grew as rapidly as -pY from lag to logarithmic and finally to stationary phases. Consequently, the metabolic burden could not account for the observed attenuation by pHcfaAC and pHcfaACE (). Thus, mechanisms other than metabolic burden might be involved in the Salmonella attenuation.
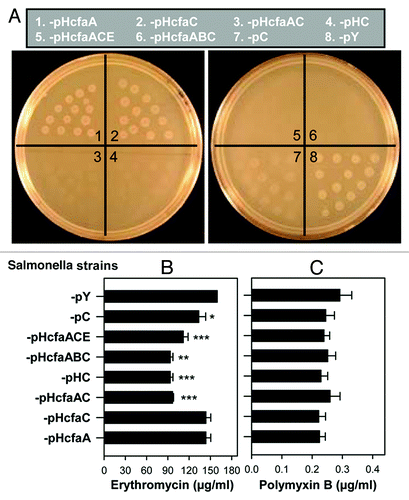
To further identify the mechanism responsible for the coordinated attenuation conferred by CfaC and CfaA upon Salmonella (), we hypothesized that these two proteins interacted, possibly forming a channel, to enable secretion of CfaB and CfaE to form fimbriae, which in turn attenuated the salmonellae. CfaA, a periplasmic chaperone, shuttles CfaB subunits to the outer cellular membrane, while CfaC, an outer membrane usher protein, orchestrates ordered tip to base assembly of fimbriae.Citation12 Neither of these two genes, when expressed alone, succeeded to attenuate Salmonella; rather, their co-expression did (). To test this hypothesis, the seven strains, P1-pHcfaA, -pHcfaC, -pHcfaAC, -pHcfaABC, -pHcfaACE, -pHC and -pY, together with the low CFA/I expression strain -pC, were tested for sensitivity to erythromycin. This 741 Dal antibiotic does not efficiently cross the outer membrane unless there is a breach of the outer membrane integrity.Citation27 The minimum inhibitory concentration (MIC) of erythromycin for strains P1-pHcfaA and -pHcfaC showed no differences when compared with control -pY (). However, the MICs for P1-pHcfaAC, -pHC, -pHcfaABC and -pHcfaACE were significantly lower than -pY (p < 0.01). These data suggested that CfaC permeability depended on chaperone CfaA, with the functional usher CfaC possibly facilitating erythromycin diffusion for uptake by salmonellae, resulting in growth inhibition. The MIC for strain P1-pC was lower than that of -pY (p < 0.05), indicating a positive correlation between CFA/I fimbriae expression level and cell permeability. The MICs for the eight strains were further determined using another membrane permeabilizer, polymyxin B, since polymyxin B also has detrimental effects on compromised bacterial outer membranes.Citation28 No differences in the polymyxin B MICs for any of these strains were found (), suggesting their outer membrane integrity remained intact after the CFA/I channel installation.
Figure 5. CfaA and CfaC together facilitate erythromycin uptake but do not lead to enhanced sensitivity to polymyxin B. Eight strains, (1) P1-pHcfaA, (2) -pHcfaC, (3) -pHcfaAC, (4) -pHC, (5) -pHcfaABC, (6) -pHcfaACE, (7) -pC and (8) -pY, were analyzed for their sensitivity to erythromycin and polymyxin B. (A) The eight strains at 105 CFUs/20 µl were used to inoculate LB agar plate containing 128 µg/ml erythromycin, incubated at 37°C and allowed to grow overnight. Depicted are representative images from one of three experiments. (B and C) Each of the eight strains was inoculated at ~4.5 × 103 CFUs/ml to liquid LB media containing various concentrations of (B) erythromycin or (C) polymyxin B. Following 37°C overnight incubation, growth or no growth was determined to obtain MICs. Differences in MICs were calculated using Tukey Kramer multiple comparisons test. Values depict the mean ± SEM of three independent experiments. *p < 0.05, **p < 0.01 and ***p < 0.001 depict significant differences of P1-pHcfaAC, -pHC, -pHcfaABC, -pHcfaACE and -pC vs control -pY.
Evaluation of protective efficacy of channel-attenuated Salmonella strains
To determine which combination of CfaA- and CfaC-bearing strains can act as a live vaccine, BALB/c mice were orally immunized, and those surviving vaccination were revaccinated with one of the four attenuated strains (). Eight weeks after the second immunization, mice were then challenged with wt H71. The best of the tested vaccines was P1-pHcfaACE, which showed 88.9% surival (p < 0.001; ). The P1-pHcfaABC vaccine was the least protective of the four vaccines, only conferring 30% protection, but was still better than sPBS-dosed control mice. This poor performance may be in part explained by its rapid clearance, which may impair its ability to stimulate protective immunity. While the co-expression of CfaA and CfaC by P1-pHcfaAC showed reduced efficacy by conferring only 60% protection, much like -pHC, the co-expression of CfaE may be important to enhance protective capacity of this vaccine.
Figure 6. Usher-attenuated P1-pHcfaACE and -pHcfaAC vaccines are protective against wt Salmonella challenge. (A) Four attenuated strains, P1-pHC (n = 10), -pHcfaABC (n = 15), -pHcfaACE (n = 10) and -pHcfaAC (n = 15), were used to orally immunize BALB/c mice, as done in ; sPBS-dosed mice served as a control. Six weeks after the second immunization, mice were orally challenged with 5 × 107 CFUs of wt H71. ***p < 0.001 depicts the survival fractions when compared with challenged, PBS-dosed mice. The data depict the mean of 2 to 3 experiments. (B) The same four attenuated strains used in (A) were used to orally immunize C57BL/6 mice, with sPBS-dosed mice as a control. Four weeks post-immunization, mice were orally challenged with 5 × 107 CFUs of wt H71. *p < 0.05 and **p < 0.01 depict the survival fractions from two experiments when compared with challenged, PBS-dosed mice. The Kaplan-Meier method was used to obtain the survival fractions after challenge for both (A) and (B) experiments.
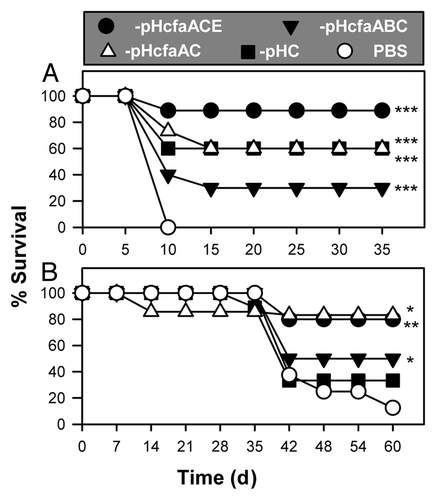
A second study was conducted to determine the efficacy of these live vaccines in C57BL/6 mice. Mice were given a single oral dose of vaccine and challenged 4 weeks later with wt H71. One of the P1-pHcfaAC-dosed mice succumbed to vaccination, but 85.7% of the mice (or 75% of the remaining mice) survived (). As with the BALB/c mice, similar vaccine efficacy was conferred by P1-pHcfaACE in C57BL/6 mice with 80% survival. P1-pHcfaABC and -pHC only achieved 50% and 33.3% survival, respectively. Collectively, these results show that making Salmonella’s outer membrane semi-permeable can attenuate its wt phenotype, and components of the cfa/I operon were valuable for conferring optimal protection in both BALB/c mice and C57BL/6 mice. Co-expression of cfaB with the usher gene dampened the protective capacity of the vaccine, possibly, because it was rapidly cleared, lessening its capacity to stimulate a protective immune response. Thus, these studies demonstrate it is possible to attenuate wt Gram-negative bacteria by making the bacilli permeable to their environment, which offers a different method to induce attenuations for use as a live vaccine.
Discussion
Although heat-labile and heat-stable enterotoxins are the major virulence factors of ETEC responsible for diarrhea in infected hosts,Citation29 CFA/I fimbriae aid in the initial infection to promote ETEC adherence to host intestinal epithelium,Citation30 thus allowing elicitation of the toxins in close proximity to the intestinal epithelium.Citation30 In fact, Abs induced to CFA/I fimbriae are protective against ETEC.Citation29,Citation31,Citation32 To enable vaccination, overexpression of CFA/I fimbriae by an attenuated Salmonella vaccine vector elicits elevated mucosal and systemic Abs to the fimbriae and also alters host recognition of the salmonellae by stimulating both Th1 and Th2 cells to the expressed fimbriaeCitation15 and salmonellaeCitation25 and dampening macrophage inflammatory responses.Citation17 In addition to these findings, the evidence presented here suggests that interfering with salmonellae permeability to their environment by cfa/I operon can promote their clearance and thus could possibly be used as a general strategy for vaccine development. As such, the data showed that varying the expression of cfa/I components impacts the level of attenuation upon wt Salmonella. Simply expressing the CFA/I fimbriae was insufficient, as evidenced by the P1-pC that remained virulent in macrophages and in mice. Thus, to enhance the expression of cfa/I operon, a strategy commonly used for eukaryotic promoters was adopted, whereby three different prokaryotic promoters were fused in order to enhance transcription and ultimately the amount of fimbrial proteins produced in the Salmonella host. The introduction of a tripartite promoter to enhance cfa/I operon expression succeeded, as evidenced by the enhanced production of CfaB by P1-pHC, its attenuation in macrophages and its ability to serve as a vaccine for salmonellosis. Such an approach in itself is novel and significantly augmented AGE to attenuate Salmonella.
To learn which of the genes in the cfa/I operon were relevant to the observed Salmonella attenuation, subsequent studies developed additional strains to test if individual genes or their combination were important for the observed attenuation. Since CfaC was identified as an usher proteinCitation12,Citation14 to allow exportation of the fimbrial gene products, CfaB encoding the major subunit and CfaE, encoding the tip protein, we hypothesized that CfaC weakens the structural integrity of the Salmonella membranes as other chaperone-usher combinations do for pilus assembly.Citation33,Citation34 As such, usher proteins typically facilitate pilus subunit translocation outside the outer membrane,Citation33 and in the case of uropathogenic E. coli, its PapC usher can form a pore in the outer membrane.Citation33,Citation34 Whether or not CfaC can form a similar pore is unclear, but it does mediate the ordered translocation of fimbrial subunits, as evident with other ETEC.Citation12 Thus, it was anticipated the expression of CfaC alone would be sufficient to attenuate Salmonella, but this was found not to be the case, as P1-pHcfaC retained its virulence in macrophages and mice. Moreover, it was resistant to erythromycin treatment, suggesting that simply expressing CfaC is insufficient for permeabilization. In fact, co-expression with CfaA by the P1-pHcfaAC strain was required for susceptibility to erythromycin treatment and loss of virulence in macrophages and in mice, which suggests usher CfaC transporting to and installing in cell outer membrane require chaperone CfaA or at least possible activation of CfaC by CfaA. Since CfaC usher is able to facilitate the secretion of large protein molecules CfaB and CfaE for fimbria formation, it is justified to conclude that excessive CfaC expression may facilitate small molecule entry to or exit from the cell freely, much like a channel or pore, as suggested by the erythromycin sensitivity studies. However, CfaC is presumed to be a dedicated channel for CFA/I fimbrial assembly and perhaps erythromycin can use CfaC to facilitate its entry into salmonellae.
Alternatively, at present, it is unclear whether expression of CfaC alters the outer membrane integrity, thus, enabling the enhanced sensitivity (uptake) of erythromycin. However, from the sensitivity study with another antibiotic polymyxin B, it was evident that the salmonellae outer membrane was not disrupted due to the co-expression of CfaAC, suggesting the outer membrane integrity remains intact. Hence, the enhanced permeability to erythromycin by the channeled strains may result from erythromycin’s use of the expressed channels to facilitate entry into the salmonellae. Regardless, co-expression of CfaAC clearly enhances the permeability of the salmonellae to erythromycin, which is in agreement with the previous observation that overexpression of curli or fimbriae can cause the bacteria to be more sensitive to erythromycin.Citation27 Studies are currently being pursued to further investigate this issue.
As a vaccine, P1-pHcfaAC was able to confer protection against wt Salmonella challenge for both BALB/c and C57BL/6 mice. For BALB/c mice, it was as protective as P1-pHC was for C57BL/6 mice and it was capable of conferring 83.7% survival as our best construct, P1-pHcfaACE. In fact, P1-pHcfaACE also conferred the best protection against wt Salmonella challenge for 88.9% survival by the vaccinated BALB/c mice. These data show the inclusion of CfaE is important for optimal protection. CFA/I fimbriae act as a hemagglutinin in which CfaB displays glycosphingolipid-binding capacity, and CfaE contributes to erythrocyte binding and is important for orchestrating the polymerization of the pilus.Citation12,Citation35 How the inclusion of CfaE in P1-pHcfaACE is important for protection against wt Salmonella challenge is unclear, although 88.9% of the challenged mice survived, unlike the mice vaccinated with the intact cfa/I operon -pHC or with -pHcfaAC, which showed a reduced efficacy. The inclusion of CfaB with CfaA and CfaC greatly attenuated Salmonella (P1-pHcfaABC), but because of such attenuation, its ability to protect was less than ideal, as evidenced by the reduced efficacy (≤ 50%) in both BALB/c and C57BL/6 mice. Furthermore, since P1-pHC conferred a similar protection as the conventional vaccine H647, this evidence suggests the described AGE method for generating live attenuated vaccines by overexpression of chaperone and usher proteins is as efficient as conventional virulence mutagenesis strategies involving the deletion of virulence genes.
Conventional methods to develop live vaccines often require prior knowledge of virulence factors. A significant attribute of the described study is the potential to express foreign antigens to attenuate Gram-negative pathogens. Simply expressing the cfa/I operon, as done for P1-pC, was found to be insufficient by wt Salmonella despite fimbria formation. Instead, elevated expression was achieved by utilizing the tripartite promoter to enhance expression of cfa/I genes as a potential vaccine candidate for salmonellosis. The P1-pHC could not colonize the mouse spleen nor could -pHcfaABC, a phenomenon only previously observed with two Salmonella ser Enteritidis mutants,Citation24 the phoP and rfaC, at a dose of 1 × 104 CFUs. However, in our study, neither P1-pHC nor -pHcfaABC could colonize mouse spleens when mice were orally dosed with 1 × 109 CFUs. Instead, these results show that overexpression of cfa/I machinery to export fimbrial subunits represents a feasible alternative to conventional attenuation methods and perhaps requires the inclusion of the CfaE fimbrial subunit for optimal attenuation of Salmonella. While the inclusion of CfaB was less effective in stimulating protective immunity against Salmonella, it was, however, very potent when combined with CfaA and CfaC in attenuating Salmonella, which implicates its possible use for other pathogens. As such, the combination effective for eliciting protective immunity to Salmonella may not be the same combination of cfa/I genes needed to attenuate other pathogens. These findings complement those of a previous study of overexpressing a Mycobacterium tuberculosis heat-shock protein found also to attenuate the bacilli, favoring enhanced immune stimulation and inducing IFN-γ-producing CD8+ T cells.Citation36
In summary, these studies suggest AGE or overexpression of chaperones and/or usher proteins may provide an alternative to attenuate virulent pathogens, either by making the pathogen more permeable to host environmental stresses or by enhancing the pathogen’s immunogenicity. Overexpression of CfaA, CfaC and CfaE represented the best combination of cfa/I to attenuate Salmonella, allowing it to be used as a live vaccine for salmonellosis. Although co-expression of CfaA, CfaB and CfaE or the entire cfa/I operon exhibited the strongest attenuation upon Salmonella, such attenuation resulted in its rapid clearance from the host disallowing optimal immunization. From this study, we expect other related heterologous proteins, such as porins and appendage structures, may also exert similar attenuating capacity upon Gram-negative bacteria.
Experimental Procedures
Mouse care and study
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and complied with the Animal Welfare Act and all other applicable federal, state and local laws. All animal care and procedures were approved by the Montana State University (MSU) Institutional Animal Care and Use Committee. Pathogen-free female BALB/c and C57BL/6N mice (National Cancer Institute, Frederick Cancer Research Facility) 7–9 weeks of age were used throughout this study. All mice were maintained at MSU Animal Resource Center under pathogen-free conditions in individually ventilated cages under HEPA-filtered barrier conditions and were fed sterile food and water ad libitum.
Bacterial strains, plasmids, media, genetic manipulation and protein gel blot
The bacterial strains, plasmids and their relevant characteristics are provided in Table S1. E. coli strain S17–1(λpir) was used for replicating R6K original vector pCVD442 and its derivative plasmids when preparing the suicide plasmid for Salmonella. The P1 (Δasd::kanR S. typhimurium H71) mutant was derived from wt strain H71. Strains containing plasmids were grown at 37°C in Lysogeny broth (LB; 10 g of tryptone, 10 g of NaCl and 5 g of yeast extract/L) with appropriate antibiotics, if needed. For S. typhimurium asd mutant construction, ampicillin (100 µg/ml) and kanamycin (10 µg/ml) were used for mutant selection. Diaminopimelic acid (DAP) (50 µg/ml) was used for E. coli H681Citation22 or S. typhimurium H71 culture unless a plasmid containing asd was introduced. Bacteria were cultured in LB and stored at -80°C in LB plus 20% glycerol.
For analyzing the growth rates of the recombinant Salmonella strains, bacteria were inoculated from -80°C freezer onto LB agar for overnight incubation at 37°C. Salmonella organisms were harvested from LB agar and then inoculated into LB liquid media with the initial OD600 adjusted to ~0.1. Inoculants were allowed to grow at 37°C in BioScreen C (Lab Systems) with agitation at 150 rpm for 4.5 h and the OD600 values were measured every 30 min. Data were downloaded and utilized for statistical comparison among these strains.
Restriction endonucleases, T4 DNA ligase and other enzymes were purchased from New England BioLabs, unless otherwise noted. Chemicals were purchased from Sigma-Aldrich. Genetic manipulations were conducted by using standard methods, as described elsewhere.Citation37 Plasmid DNA was extracted by using a Qiagen Miniprep Kit. DNA fragments were purified and extracted from agarose gel slices, using Qiagen QIAquick Gel Extraction Kit. Competent E. coli and S. typhimurium cells were made in 10% glycerol and transformed with electroporation.
To verify the expression level of CFA/I in P1-pHC and -pC, 6 × 108 CFUs of P1-pHC, -pC, or -pY whole cells harvested from liquid LB cultures were boiled with SDS-PAGE loading buffer at 95°C and loaded into a 12% [wt/vol] SDS-PAGE gel. Proteins were transferred to 0.2-µm-pore-size nitrocellulose membranes (Pall Corporation). Membranes were probed first with polyclonal anti-CFA/I rabbit sera and then with goat anti-rabbit IgG conjugated to horseradish peroxidase (Southern Biotechnology Associates, Inc.). Detection of CfaB was achieved upon development with the substrate 4-chloro-1-naphthol chromogen and H2O2 (Sigma Chemical Co.), and the expression levels of CfaB from P1-pHC and -pC were compared. The protein marker used was Prestained Precision Plus Protein Standards (Bio-Rad Labs).
Construction of mutant, plasmids and recombinant strains
The P1 mutant was generated by integrating the recombinant suicide plasmid pCVDasd::kanR (Table S1), which was constructed as follows: Two polymerase chain reaction (PCR) DNA fragments, both upstream (1757 bp) and downstream (1889 bp) of H71 asd flanking the 1005 bp inner region of asd, were fused together via two cloning steps. The two pairs of primers, asd-dn-F/asd-dn-R and asd-up-F/asd-up-R (Table S2), used for amplifying the upstream and downstream DNA fragments of H71 asd, were designed according to the genomic sequence of S. typhimurium LT2 (Accession number NC_003197)Citation38 since H71 genome sequence was not available. These two fragments were ligated together by introducing an XbaI site in the junction. Meanwhile, a kanamycin resistance gene coding cassette (kanR) from pCR2.1-TOPO (Invitrogen) was amplified using a pair of primers of kan-F/kan-R with XbaI integrated at both of their 5′ ends (Table S2). The kanR DNA fragment was digested with XbaI and inserted between upstream and downstream of asd DNA fragments at the XbaI junction site to form a sandwich DNA fragment. Next, this sandwich DNA fragment was subcloned to suicide vector pCVD442 (ampR) between SalI and SacI. After transforming ligates to E. coli S17–1 and selecting on an LB plate containing ampicillin (100 µg/ml) and kanamycin (10 µg/ml), an asd suicide plasmid was generated and termed pCVDasd::kanR. pCVDasd::kanR was transformed to H71. Transformants were selected on LB agar containing 10 µg/ml kanamycin and incubated at 37°C overnight. A single colony was picked and grown to late logarithmic phase in LB with 50 µg/ml DAP. A series of dilutions was plated on LB agar containing no NaCl, but 5% sucrose and 50 µg/ml DAP. Sucrose-resistant colonies were tested for loss of asd by both kanamycin resistance testing and the obligated requirement for DAP. Those colonies that did not grow without DAP, but were resistant to kanamycin, were thereby asd mutants. A total of three of these asd mutants were obtained. One clone was further analyzed by sequencing to confirm its asd inner sequence was replaced by kanR.
To enhance cfa/I operon expression, the macrophage inducible promoters, PphoP and PpagC, were fused with promoter PtetA from plasmid pC (Table S1) and installed upstream of the cfa/I operon, as follows: The 274 bp promoter PpagC was amplified from template H71 genomic DNA by PCR with a pair of primers, pagC-F/pagC-R (Table S1) and was inserted between PphoP and PtetA in plasmid pV55.Citation39 This construct was termed pV6 (Table S1). Then cfa/I operon from pC (Table S1) was used to replace lcrV in pV6. The new plasmid in which the cfa/I operon was controlled by fused promoter, PtetA~PpagC~PphoP, was termed pHC.
Construction of CFA/I non-fimbriae genes cfaA and cfaC expression plasmids
To determine if the non-fimbriae genes cfaA and cfaC were involved in the Salmonella attenuation, the inner DNA fragment sequences of the cfaB encoding fimbrial major subunit and cfaE encoding fimbrial minor subunit from plasmid pHC were in-frame deleted or completely removed, respectively. The primers used for cfaB and cfaE deletion were cfaA1-F/cfaB-R and cfaE-F/cfaE-R, respectively (Table S2). After deletion, the upstream sequence of cfaB had 21 bp and downstream had 63 bp left, which were confirmed by sequencing. This cfaB in-frame deletion plasmid derived from pHC was named pHcfaACE. The cfaE was completely deleted from the pHC, and the new plasmid was named pHcfaABC. The double mutant of cfaBE was derived from pHcfaABC, whereby both cfaA and cfaC expression were under the regulation of fusion promoter. This mutant was termed pHcfaAC. The plasmid pHV was generated by digesting pHC with SacI and XhoI and treated with T4 DNA polymerase and self-ligated. Thus, pHV harbors PtetA~PpagC~PphoP fusion promoter, but not the cfa/I component. Plasmid pY containing only asd with its replication origin was derived from pC by digesting pC with ScaI and then self-ligated.
To determine the involvement of chaperone gene cfaA in the Salmonella attenuation, cfaA was amplified by PCR with a pair of primers of cfaA2-F/cfaA-R (Table S2) and cloned downstream of the fusion promoter of pHC. Thus, cfaABCE in pHC was replaced by cfaA. After sequencing confirmation, this new plasmid was named pHcfaA. To determine the involvement of the usher gene cfaC in the Salmonella attenuation, the pHcfaAC were digested by BamHI and SacI to remove the upstream genes cfaA and in-framed deleted cfaB, and the sticky ends were filled by T4 DNA polymerase and were self-ligated. This plasmid was termed pHcfaC.
AFM studies
AFM has been successfully used to visualize bacterial pili and fimbriae.Citation40 In this study, CFA/I fimbriae expression was detected via AFM for P1-pHC, -pC, -pY and all the other cfa/I mutants, e.g., P1-pHcfaABC, -pHcfaACE, -pHcfaAC, -pHcfaC and pHcfaA. Details of the sample preparation for AFM imaging were previously reported.Citation41 Briefly, bacterial cells were grown in liquid LB medium and shaken at 75 rpm at 37°C until their OD600 reached ~0.5. One hundred microliters of each culture suspension was directly dropped onto a fresh cleaved mica disk, and the suspension covered mica was kept for 10 min at room temperature, followed by rinsing with sPBS or Nanopure water. This sample was then dried with a nitrogen gas flow. All AFM images were acquired in air with a Nanoscope V Extended Multimode system from Veeco. Imaging was performed in tapping mode to reduce the tip sample interaction and the lateral forces. Two types of silicon probes were used for AFM experiments, namely, RTESPW from Veecoprobes and NSC18 from MikroMasch USA. At least 20 cells were observed for each strain. Tapping mode images were shown since more morphological details of bacterial cells can be revealed in tapping mode images than height images.Citation41
Evaluation of infection and replication of recombinant Salmonella strains in RAW264.7 macrophages
RAW264.7 macrophages (American Type Culture Collection) were used for evaluating strains P1-pHC, -pC and -pY infection and replication. Infections were conducted exactly as previously described.Citation17 1.25 × 106 RAW264.7 cells/well without antibiotics were allowed to adhere to plastic in 24-well microtiter dishes (B-D Labware) at 37°C with 5% CO2. Wells were washed and the nonadherent cells were collected and counted to determine cell numbers that remained plastic-adherent. After overnight culture, cells were infected with varying bacteria to macrophage ratios, 1:1, 10:1 and 100:1, for 1 h at 37°C. Wells were washed twice and then incubated with 50 µg/ml of gentamicin for 30 min at 37°C. After washing twice, as described, fresh medium without antibiotics was added and cells were incubated for either an additional 4 or 24 h. Next, the macrophages were water lysed and the bacteria were used to make a series of dilutions on LB agar plates. After incubation overnight at 37°C, the bacterial CFUs were determined. The cfa/I mutants recombined strains of P1-pHcfaACE, -pHcfaABC, -pHcfaAC, -pHcfaA and -pHcfaC with control -pY were used to infect RAW264.7 cells, as stated above, while a bacteria to macrophage ratio of 1:1 was adopted.
Assessment of sensitivity of recombinant Salmonella to antibiotics
Strains P1-pHcfaA, -pHcfaC, -pHcfaAC, -pHC, -pHcfaACE, -pHcfaABC and -pC with control -pY were tested for sensitivity to serial dilutions of erythromycin and polymyxin B beginning at 508 µg/ml and 10 µg/ml, respectively, in liquid LB media. The MIC was determined according to standard method.Citation42 This experiment was used to verify outer membrane permeability to erythromycin and polymyxin B.
Mouse studies
To assess the virulence of the newly constructed P1-based strains, groups of BALB/c mice (5–8/group; experiments were performed 2 or 3 times) were orally gavaged with 1 × 109 CFUs of P1-pHC, -pC, -pY, -pHcfaA, -pHcfaC, -pHcfaAC, -pHcfaACE or -pHcfaABC, or orally gavaged with 5 × 109 ΔaroA S. typhimurium vaccine strain H647Citation15,Citation20 in 200 µl sPBS and compared with virulence in mice orally gavaged with 1 × 109 CFUs of the wt S. typhimurium strain H71 (50% lethal dose, 5 × 104 CFUs).Citation20 To assess sensitivity to the vaccines, groups were monitored for survival for 3–6 weeks post-infection. To assess the extent of inflammation and tissue colonization, spleens, PPs and livers were evaluated at 5 d post-infection. Splenic weights were determined to measure the extent of inflammation. Spleens, PPs and livers were then dounce homogenized and assayed for extent of tissue colonization. Serial dilutions were made and samples incubated at 37°C on LB agar. The bacterial burden in tissues is expressed as total CFUs per tissue.
Serum IgG and fecal IgA endpoint Ab titers were determined by standard ELISA methods.Citation16 Serum and fecal samples were collected every two weeks from individual mice infected with one of the described Salmonella-based vaccines and each mouse was assessed for reactivity to purified CFA/I fimbriae and heat-killed S. typhimurium (HKST). Wild-type H71 strain was used as source of HKST inactivated by incubating live cells at 100°C for 15 min and absence of cell growth was confirmed on LB agar. Fecal pellets were collected from individual mice and solubilized in 50 µg/ml of soybean trypsin inhibitor (Sigma-Aldrich) in sPBS (10% v/w) by continual vortexing for 30 min at 4°C. After microcentrifugation, supernatants were frozen until assayed. Flat-bottom, 96-wells (MaxiSorb, Nunc) were coated with CFA/I fimbriae (5 μg/ml) or 2.5 × 108 CFUs HKST and incubated at 4°C overnight, followed by standard ELISA protocols.Citation16 Bound serum IgG and mucosal IgA Abs were detected with horseradish peroxidase-conjugated goat anti-mouse IgG or IgA (South. Biotech. Assoc.) and developed in the presence of enzyme substrate, 2,2′-azinobis (3-ethylbenthiazoline-6-sulfonic acid; Moss Inc.). The endpoint titers were determined as the reciprocal dilutions of the last dilution yielding an absorbance at OD415 above 0.100 OD units above negative controls.
For the challenge studies, BALB/c mice were orally immunized twice with 1 × 109 CFUs of vaccine at 4 week intervals and C57BL/6N mice were immunized with a single oral dose of vaccine. Mice were orally challenged with 5 × 107 CFUs of wt S. typhimurium H71 in 200 µl sPBS 4 weeks after the last immunization. The amount of bacteria given was confirmed by plating serial dilutions of bacterial suspensions onto LB agar plates. Mice were observed twice daily and the extent of survival was recorded for 4 weeks.
Statistical analysis
The differences among the bacterial CFUs and tissue weights were calculated by Tukey Kramer multiple comparisons test. The Kaplan-Meier method (GraphPad Prism, GraphPad Software, Inc.) was applied to obtain the survival fractions following infection with a lethal dose of wt S. typhimurium H71. Using the Mantel-Haenszel log rank test, the p-value for statistical differences between vehicle and vaccines was discerned at the 95% confidence interval. The Student's t-test (GraphPad Prism, GraphPad Software, Inc.) was used for calculating the difference between ELISA titers.
| Abbreviations: | ||
| Ab | = | antibody |
| AFM | = | atomic force microscopy |
| AGE | = | attenuating gene expression |
| CFA/I | = | colonization factor antigen I |
| DAP | = | diaminopimelic acid |
| ETEC | = | enterotoxigenic E. coli |
| MIC | = | minimum inhibitory concentration |
| PP | = | Peyer’s patches |
| wt | = | wild-type |
Additional material
Download Zip (331.7 KB)Acknowledgments
The authors thank Drs. Jerod Skyberg and Massimo Maddaloni for their helpful comments and Ms. Nancy Kommers for her assistance in preparing this manuscript. This work is supported by grants from National Institutes of Health Grant R21 AI-080960, R01 AI-41123, P20 RR020185, an equipment grant from the M.J. Murdock Charitable Trust, Montana Agricultural Experiment Station and US Department of Agriculture Formula Funds and Office of Navy Research Award N00014-10-1-0946.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Ly KT, Casanova JE. Mechanisms of Salmonella entry into host cells. Cell Microbiol 2007; 9:2103 - 11; http://dx.doi.org/10.1111/j.1462-5822.2007.00992.x; PMID: 17593246
- Ohl ME, Miller SI. Salmonella: A model for bacterial pathogenesis. Annu Rev Med 2001; 52:259 - 74; http://dx.doi.org/10.1146/annurev.med.52.1.259; PMID: 11160778
- Arrach N, Porwollik S, Cheng P, Cho A, Long F, Choi SH, et al. Salmonella serovar identification using PCR-based detection of gene presence and absence. J Clin Microbiol 2008; 46:2581 - 9; http://dx.doi.org/10.1128/JCM.02147-07; PMID: 18524964
- Pang T, Levine MM, Ivanoff B, Wain J, Finlay BB. Typhoid fever - important issues still remain. Trends Microbiol 1998; 6:131 - 3; http://dx.doi.org/10.1016/S0966-842X(98)01236-0; PMID: 9587187
- Mulvey MR, Boyd DA, Olson AB, Doublet B, Cloeckaert A. The genetics of Salmonella genomic island 1. Microbes Infect 2006; 8:1915 - 22; http://dx.doi.org/10.1016/j.micinf.2005.12.028; PMID: 16713724
- Chan K, Kim CC, Falkow S. Microarray-based detection of Salmonella enterica serovar Typhimurium transposon mutants that cannot survive in macrophages and mice. Infect Immun 2005; 73:5438 - 49; http://dx.doi.org/10.1128/IAI.73.9.5438-5449.2005; PMID: 16113260
- Hormaeche CE, Joysey HS, Desilva L, Izhar M, Stocker BA. Immunity conferred by Aro- Salmonella live vaccines. Microb Pathog 1991; 10:149 - 58; http://dx.doi.org/10.1016/0882-4010(91)90075-L; PMID: 1890952
- Galán JE, Curtiss R 3rd. Virulence and vaccine potential of phoP mutants of Salmonella typhimurium. Microb Pathog 1989; 6:433 - 43; http://dx.doi.org/10.1016/0882-4010(89)90085-5; PMID: 2671582
- Sommerfelt H, Steinsland H, Grewal HM, Viboud GI, Bhandari N, Gaastra W, et al. Colonization factors of enterotoxigenic Escherichia coli isolated from children in north India. J Infect Dis 1996; 174:768 - 76; http://dx.doi.org/10.1093/infdis/174.4.768; PMID: 8843215
- Jonson AB, Normark S, Rhen M. Fimbriae, pili, flagella and bacterial virulence. Contrib Microbiol 2005; 12:67 - 89; http://dx.doi.org/10.1159/000081690; PMID: 15496777
- Baker KK, Levine MM, Morison J, Phillips A, Barry EM. CfaE tip mutations in enterotoxigenic Escherichia coli CFA/I fimbriae define critical human intestinal binding sites. Cell Microbiol 2009; 11:742 - 54; http://dx.doi.org/10.1111/j.1462-5822.2009.01287.x; PMID: 19207729
- Li YF, Poole S, Rasulova F, McVeigh AL, Savarino SJ, Xia D. A receptor-binding site as revealed by the crystal structure of CfaE, the colonization factor antigen I fimbrial adhesin of enterotoxigenic Escherichia coli. J Biol Chem 2007; 282:23970 - 80; http://dx.doi.org/10.1074/jbc.M700921200; PMID: 17569668
- Anantha RP, McVeigh AL, Lee LH, Agnew MK, Cassels FJ, Scott DA, et al. Evolutionary and functional relationships of colonization factor antigen I and other class 5 adhesive fimbriae of enterotoxigenic Escherichia coli. Infect Immun 2004; 72:7190 - 201; http://dx.doi.org/10.1128/IAI.72.12.7190-7201.2004; PMID: 15557644
- Saulino ET, Bullitt E, Hultgren SJ. Snapshots of usher-mediated protein secretion and ordered pilus assembly. Proc Natl Acad Sci USA 2000; 97:9240 - 5; http://dx.doi.org/10.1073/pnas.160070497; PMID: 10908657
- Pascual DW, Hone DM, Hall S, van Ginkel FW, Yamamoto M, Walters N, et al. Expression of recombinant enterotoxigenic Escherichia coli colonization factor antigen I by Salmonella typhimurium elicits a biphasic T helper cell response. Infect Immun 1999; 67:6249 - 56; PMID: 10569734
- Jun S, Gilmore W, Callis G, Rynda A, Haddad A, Pascual DW. A live diarrheal vaccine imprints a Th2 cell bias and acts as an anti-inflammatory vaccine. J Immunol 2005; 175:6733 - 40; PMID: 16272329
- Pascual DW, Trunkle T, Sura J. Fimbriated Salmonella enterica serovar Typhimurium abates initial inflammatory responses by macrophages. Infect Immun 2002; 70:4273 - 81; http://dx.doi.org/10.1128/IAI.70.8.4273-4281.2002; PMID: 12117936
- Ochoa-Repáraz J, Riccardi C, Rynda A, Jun S, Callis G, Pascual DW. Regulatory T cell vaccination without autoantigen protects against experimental autoimmune encephalomyelitis. J Immunol 2007; 178:1791 - 9; PMID: 17237429
- Kochetkova I, Trunkle T, Callis G, Pascual DW. Vaccination without autoantigen protects against collagen II-induced arthritis via immune deviation and regulatory T cells. J Immunol 2008; 181:2741 - 52; PMID: 18684965
- Walters N, Trunkle T, Sura M, Pascual DW. Enhanced immunoglobulin A response and protection against Salmonella enterica serovar Typhimurium in the absence of the substance P receptor. Infect Immun 2005; 73:317 - 24; http://dx.doi.org/10.1128/IAI.73.1.317-324.2005; PMID: 15618168
- Galán JE, Nakayama K, Curtiss R 3rd. Cloning and characterization of the asd gene of Salmonella typhimurium: use in stable maintenance of recombinant plasmids in Salmonella vaccine strains. Gene 1990; 94:29 - 35; http://dx.doi.org/10.1016/0378-1119(90)90464-3; PMID: 2227450
- Wu S, Pascual DW, VanCott JL, McGhee JR, Maneval DR Jr., Levine MM, et al. Immune responses to novel Escherichia coli and Salmonella typhimurium vectors that express colonization factor antigen I (CFA/I) of enterotoxigenic E. coli in the absence of the CFA/I positive regulator cfaR. Infect Immun 1995; 63:4933 - 8; PMID: 7591160
- Suo Z, Avci R, Yang X, Pascual DW. Efficient immobilization and patterning of live bacterial cells. Langmuir 2008; 24:4161 - 7; http://dx.doi.org/10.1021/la7038653; PMID: 18321142
- Karasova D, Sebkova A, Vrbas V, Havlickova H, Sisak F, Rychlik I. Comparative analysis of Salmonella enterica serovar Enteritidis mutants with a vaccine potential. Vaccine 2009; 27:5265 - 70; http://dx.doi.org/10.1016/j.vaccine.2009.06.060; PMID: 19577637
- Pascual DW, White MD, Larson T, Walters N. Impaired mucosal immunity in L-selectin-deficient mice orally immunized with a Salmonella vaccine vector. J Immunol 2001; 167:407 - 15; PMID: 11418677
- Glick BR. Metabolic load and heterologous gene expression. Biotechnol Adv 1995; 13:247 - 61; http://dx.doi.org/10.1016/0734-9750(95)00004-A; PMID: 14537822
- Robinson LS, Ashman EM, Hultgren SJ, Chapman MR. Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol Microbiol 2006; 59:870 - 81; http://dx.doi.org/10.1111/j.1365-2958.2005.04997.x; PMID: 16420357
- Sikora AE, Lybarger SR, Sandkvist M. Compromised outer membrane integrity in Vibrio cholerae Type II secretion mutants. J Bacteriol 2007; 189:8484 - 95; http://dx.doi.org/10.1128/JB.00583-07; PMID: 17890307
- Sánchez J, Holmgren J. Virulence factors, pathogenesis and vaccine protection in cholera and ETEC diarrhea. Curr Opin Immunol 2005; 17:388 - 98; http://dx.doi.org/10.1016/j.coi.2005.06.007; PMID: 15963708
- Gaastra W, Svennerholm AM. Colonization factors of human enterotoxigenic Escherichia coli (ETEC). Trends Microbiol 1996; 4:444 - 52; http://dx.doi.org/10.1016/0966-842X(96)10068-8; PMID: 8950814
- Yang X, Thornburg T, Holderness K, Suo Z, Cao L, Lim T, et al. Serum antibodies protect against intraperitoneal challenge with enterotoxigenic Escherichia coli. J Biomed Biotechnol 2011; 2011:732848; http://dx.doi.org/10.1155/2011/632396; PMID: 21541193
- Boedeker EC. Vaccines for enterotoxigenic Escherichia coli: current status. Curr Opin Gastroenterol 2005; 21:15 - 9; PMID: 15687879
- Waksman G, Hultgren SJ. Structural biology of the chaperone-usher pathway of pilus biogenesis. Nat Rev Microbiol 2009; 7:765 - 74; http://dx.doi.org/10.1038/nrmicro2220; PMID: 19820722
- Remaut H, Tang C, Henderson NS, Pinkner JS, Wang T, Hultgren SJ, et al. Fiber formation across the bacterial outer membrane by the chaperone/usher pathway. Cell 2008; 133:640 - 52; http://dx.doi.org/10.1016/j.cell.2008.03.033; PMID: 18485872
- Tchesnokova V, McVeigh AL, Kidd B, Yakovenko O, Thomas WE, Sokurenko EV, et al. Shear-enhanced binding of intestinal colonization factor antigen I of enterotoxigenic Escherichia coli. Mol Microbiol 2010; 76:489 - 502; http://dx.doi.org/10.1111/j.1365-2958.2010.07116.x; PMID: 20345656
- Stewart GR, Snewin VA, Walzl G, Hussell T, Tormay P, O'Gaora P, et al. Overexpression of heat-shock proteins reduces survival of Mycobacterium tuberculosis in the chronic phase of infection. Nat Med 2001; 7:732 - 7; http://dx.doi.org/10.1038/89113; PMID: 11385512
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2d ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press,1989.
- McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 2001; 413:852 - 6; http://dx.doi.org/10.1038/35101614; PMID: 11677609
- Yang X, Hinnebusch BJ, Trunkle T, Bosio CM, Suo ZY, Tighe M, et al. Oral vaccination with Salmonella simultaneously expressing Yersinia pestis F1 and V antigens protects against bubonic and pneumonic plague. J Immunol 2007; 178:1059 - 67; PMID: 17202369
- Touhami A, Jericho MH, Boyd JM, Beveridge TJ. Nanoscale characterization and determination of adhesion forces of Pseudomonas aeruginosa pili by using atomic force microscopy. J Bacteriol 2006; 188:370 - 7; http://dx.doi.org/10.1128/JB.188.2.370-377.2006; PMID: 16385026
- Suo Z, Yang X, Avci R, Kellerman L, Pascual DW, Fries M, et al. HEPES-stabilized encapsulation of Salmonella typhimurium. Langmuir 2007; 23:1365 - 74; http://dx.doi.org/10.1021/la0621721; PMID: 17241060
- Prouty AM, Van Velkinburgh JC, Gunn JS. Salmonella enterica serovar Typhimurium resistance to bile: identification and characterization of the tolQRA cluster. J Bacteriol 2002; 184:1270 - 6; http://dx.doi.org/10.1128/JB.184.5.1270-1276.2002; PMID: 11844755