Abstract
Currently, intercellular chemical signaling in bacteria, known as quorum sensing, is described for several species of bacteria; however, for many clinically important pathogens this significant sensory mechanism remains unknown. Among such pathogens are the pyogenic streptococci that include groups A and B streptococcus (GAS, GBS). Evidence now points to a family of transcription factors, known as Rgg/GadR/MutR, can serve as receptors for secreted pheromones. Within the genome of Streptococcus pyogenes four Rgg paralogs can be identified, two of which (Rgg2 and Rgg3) were shown to rely on short hydrophobic peptides (SHPs) to control transcription of their target promoters. SHPs were found to promote biofilm development and could offset biofilm-dispersion effects caused by Rgg1. Since Rgg homologs are present in genomes throughout Firmicute species, their newfound ability to serve as quorum-sensing mediators offers a potential opportunity to manipulate bacterial behaviors by interfering with communication networks.
Three decades of efforts have established the paradigm that communication between bacterial cells occurs via chemical signaling. In a process termed quorum sensing, bacteria secrete small molecules into their environment to tell their siblings and neighbors of their presence and willingness to work together. These communications coordinate a wide variety of bacterial behaviors that include the development of biofilms, the triggering of pathogenic attacks, the assault on neighbors to acquire new genetic material, and several other complex actions. While considerable knowledge of how quorum sensing operates and of the benefits it provides to communities has been gleaned from model organisms and a few pathogenic species, many important bacterial taxa seem virtually silent when it comes to intercellular communication due to lack of recognizable orthologous quorum-sensing signals or receptors. Such has been the case for many members of the large group of Gram-positive bacteria known as streptococci. Many species of this phylum, including groups A and B streptococcus (GAS, S. pyogenes and GBS, S. agalactiae, respectively) largely lack proteins that provide these features.
Recent findings, however, reveal a new purpose for a widespread family of proteins among Gram-positive bacteria enabling the cellular equivalent of social networking. The family of proteins known as Rgg/GadR/MutR is found throughout most Firmicute species, and though its members are known to serve as transcription factors, clearly containing a recognizable DNA-binding helix-turn-helix motif and being necessary for transcriptional activation of many genes among the streptococci, little has been explained for how they differentially control target gene expression. Despite lacking recognizable primary-sequence similarity to any other quorum-sensing components, structural prediction algorithms reveal potentially similar secondary and tertiary structure to PlcR and PrgX, two prototypical members of the Rap/NprR/PrgX/PlcR (RNPP) protein family, also found throughout Gram-positive bacteria. Each member of this family serves as a receptor for imported signaling peptides. Upon ligand interaction, RNPP proteins respond with changes in their regulatory activity.
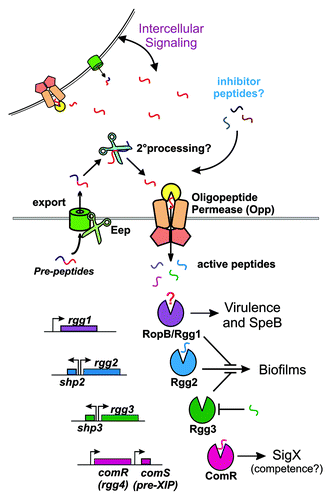
All currently available genome sequences of S. pyogenes (all coming from clinical isolates attributable to various diseases) each contain four rgg paralogs. The best studied is RopB (Rgg1), a transcription factor known for its requirement as an activator of the secreted cysteine protease SpeB. SpeB is among the most important GAS virulence factors. Another Rgg protein is ComR, found recently to control competence development in Streptococcus mutans and Streptococcus thermophilus. Adjacent to comR in these genomes is comS, a small gene encoding a peptide pheromone that interacts directly with ComR. In S. mutans, the pheromone, called XIP, converts ComR into an active transcription factor, thereby inducing expression of the alternative sigma factor, SigX. Our unpublished results indicate ComR and ComS are capable of inducing sigX expression in GAS as well.
These findings provided the first proof of principle that an Rgg protein could respond to a peptide pheromone and led to the question: are all Rgg proteins peptide receptors? Two uncharacterized rgg genes in GAS (rgg2 and rgg3, numbered in order to their similarity to RopB), offered an exceptional choice to test this hypothesis, as both rgg genes lie adjacent to small open reading frames encoding short hydrophobic peptides (SHPs, with gene names shp2 and shp3), thus providing a direct target for mutagenesis.
Indeed, it turns out that differential transcriptional activity provided by both Rgg2 and Rgg3 relies on secreted and processed forms of the peptides. The shp genes encode 22 and 23 amino acid peptides that are processed to mature, eight C-terminal amino acid pheromones. Remarkably, bioactive SHP2 and SHP3 differ by only one hydrophobic amino acid [DI(I/L)IIVGG], yet elicit different responses in DNA-binding properties, at least for Rgg3. How are these highly similar and hydrophobic pheromones distinguished by their receptors? Genetic and structural studies may provide answers. Furthermore, given the short length of the pre-peptides and hydrophobicity of the mature pheromones, one is led to wonder how peptides of such length are targeted for secretion and whether a need exists to overcome non-specific association between the hydrophobic peptides and the cell surface. Other known pheromone pre-peptides are typically twice as long (> 35 amino acids), offering more room for a secretion signal sequence, or are processed from a substantially larger secreted protein. The smallest pre-peptides predicted to partake in Rgg signaling pathways are the putative pre-XIP peptides of S. macacae and S. bovis, which are 12 and 14 amino acids long, respectively. It is not currently known how these XIP peptides are exported from the cell.
Another surprise was the finding that Rgg2 and Rgg3 each control transcription of the promoters driving both shp genes but do so with antagonistic activities. In the absence of pheromone, Rgg3 binds to DNA at both shp promoters and represses their transcription. When pheromones are present, Rgg3 releases DNA and transcription is unblocked. Oppositely, Rgg2 appears to serve only as a transcriptional activator and is inactive until SHPs are transported into the cell. Providing either SHP to Rgg2 leads to robust induction of the target promoters. The net response to pheromones, therefore, is to boost promoter expression, and since pheromone production is increased in this process, the circuit is amplified by a positive feedback loop. The use of two transcriptional regulators to control shp promoters, when one would theoretically suffice, suggests a benefit to this higher regulatory complexity. Indications that Rgg2 and Rgg3 respond differently to each peptide points to a transiently ordered response of promoters. Ongoing studies also indicate that Rgg2 and Rgg3 bind at the same location of shp promoters, so how one promotes transcription while the other represses it remains to be seen. Analogously, PlcR, a transcriptional activator, and PrgX, a repressor, have similar structures but control transcription and respond to peptide ligands via different mechanisms. Therefore, structural studies of Rgg2 and Rgg3 may reveal new mechanisms for transcriptional regulation and ligand-receptor interactions.
What function does the Rgg2/3 pathway play in GAS biology? Reverse-genetic experiments always face the difficulty of associating gene function to physiologically-relevant biological effects. However, because quorum sensing provides a means for bacterial cells to socialize, the role of Rgg2/3 in social activities is worth investigating. Biofilm development, a common example of community behavior, remains an intriguing but poorly understood lifestyle for S. pyogenes; studies are just beginning to probe how and where GAS biofilms are established in the body. Associations have been found between GAS biofilm capacity and antibiotic treatment failure, and biofilm-like structures have been described within GAS-infected tissues. Might biofilms be under the regulatory control of quorum sensing pathways in GAS? Providing synthetic SHP pheromones to cultures of wild-type NZ131 GAS cells led to a near doubling in biofilm biomass, indicating an ability of SHPs to specifically enhance biofilm development. Rgg2, the transcriptional activator, was required to produce biofilms, as was the oligopeptide permease, Opp. Interestingly, the response to SHPs was greatly enhanced when RopB (Rgg1), the activator of SpeB protease, was deleted. SpeB is known to act upon the extracellular matrix of GAS biofilms and may facilitate cellular dispersion, but it remains to be seen if the protease also interferes with peptide signaling or if RopB causes disruption of SHP signaling via other regulatory pathways. Therefore, it appears that the Rgg2/3 circuit promotes biofilm development while a separate Rgg (RopB) prevents or disassembles biofilms. Of course the fact that RopB has served as the prototypical Rgg protein among Gram-positive bacteria deserves the question if it too relies on a peptide pheromone. Past studies would indicate this is a likely possibility seeing as though a missing growth-phase-associated factor is necessary for speB expression and opp mutants disrupt proper SpeB expression. However, to date, no clear pheromone candidates have been identified and neither SHP2, SHP3, nor XIP appear to affect RopB activity.
Significantly, these studies demonstrate that at least some streptococcal behaviors can be manipulated by applying synthetic signaling peptides to cells. Taking this idea a step further, one could imagine scenarios where the course of a streptococcal infection could be influenced by synthetic peptides designed to block innate signaling. Alternatively, using peptides to inhibit streptococcal biofilms may keep streptococci in a state that is susceptible to antimicrobial drugs, thus improving success in treating recurrent infections. Two attributes of these signaling systems that could substantially contribute to the feasibility in developing therapeutics rest on the basic findings that the peptides are unmodified and linear and are actively imported to the cytoplasm where they interact with the Rgg receptors. Since streptococcal pheromones appear to have a simple linear form, unlike the cyclical pheromones of Staphylococcus, and do not possess modified amino acids seen in many other bioactive bacterial peptides, developing peptide molecules that compete with pheromone-Rgg interactions appears to be a straightforward challenge. Technologies like peptide arrays and phage display provide libraries of molecules that could be screened for abilities to interact, and possibly interfere, with pheromone-Rgg interactions. Second, since peptides are transported into the cytoplasm indiscriminately by an oligopeptide transporter, designed inhibitors, if peptides, would not face a cell-permeability issue that is of concern when designing drugs. In this case, the inhibitory peptide would gain access to the cytoplasm the same way other peptides are brought into the cell, unbeknownst to serve as a Trojan horse.
Throughout the Firmicute phylum, homologs of Rgg proteins are present in all species of Streptococcus as well as many of the Lactobacillus, Listeria, and Enterococcus families. Interestingly, some isolates of Streptococcus pneumoniae, a species that resides in the nasopharynx along with S. pyogenes, contains nearly identical copies of rgg3 and shp3 genes. The genetic neighborhood surrounding these genes on the S. pneumoniae chromosome, however, is unlike the one nearby rgg3/shp3 in GAS, possibly suggesting that different sets of genes are influenced by SHP3 among the two organisms. Likewise, nearly identical copies of rgg2 and shp2 are found in Streptococcus agalactiae. Recognition that Rgg proteins serve as pheromone receptors not only reveal a new quorum sensing pathway in Gram-positive bacteria, but also provokes questions of whether these bacteria, commonly the most abundant in the human microbiome, take part in interspecies conversations that lead to beneficial or detrimental health outcomes of the host.
Figure 1. Proposed model of Rgg-dependent quorum sensing in Streptococcus pyogenes. Pre-peptides, encoded by shp, comS and other potential small ORFs, are secreted through a yet to be identified system, are processed to mature pheromones during export (Eep is one identified protease involved in SHP2 and SHP3 development) or once outside the cell (potential secondary processing involved), and are imported back to the cytoplasm by the oligopeptide transporter. Inside the cell, the active pheromones engage Rgg proteins, thus affecting target gene expression. Peptides that disrupt Rgg-pheromone interactions presumably would be imported in the same manner as other peptides, and may serve to inhibit normal signaling.