Figures & data
Table 1 Oncogenic human viruses and viral oncogenes.
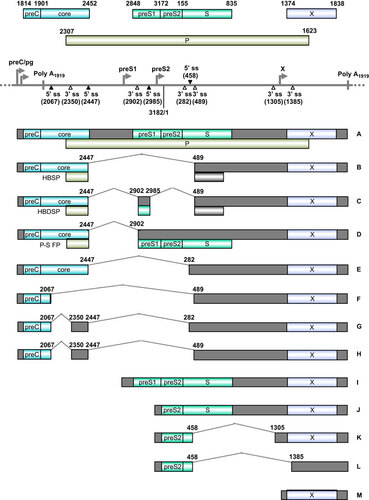
Figure 1 HPV16 E6 and E7 and alternative RNA splicing. (A) Major functional domains and motifs of the HPV16 E6, E6*I, E6ˆE7 and E7 proteins. HPV16 E6ˆE7 has the N-terminal half of E6 and the C-terminal half of E7. (B) Alternative RNA splicing products of HPV16 E6E7 pre-mRNA. Alternative RNA splicing (dashed lines) takes place from three 5′ ss at nt 191, 221 and 226 to three alternative 3′ ss at nt 409, 526 and 742.Citation27 The majority of RNA splicing occurs from the nt 226 5′ ss to the nt 409 3′ ss to produce E6*I, which is responsible for E7 translation.Citation28 E6*III derived from the nt (226 5′ ss to the nt 3358 3′ ss and E6*IV derived from the nt 226 5′ ss to the nt 2709 3' ssCitation29 are not included in this diagram. (C) Illustration of a ribosomal scanning model in HPV16 E6 and E7 translation, which is regulated by RNA splicing. Full-length E6 is translated from the unspliced E6 mRNA (upper diagram). E7 (nt 562–858) is translated from spliced E6*I mRNA in which a premature stop codon (UAA) is introduced by RNA splicing to form the E6*I ORF (lower diagram), which enlarges the space between the two ORFs. This enables a scanning ribosome to terminate translation of E6*I and reinitiate translation of E7. Nucleotide positions are numbered according to the HPV16 reference genome (PaVE, http://pave.niaid.nih.gov).
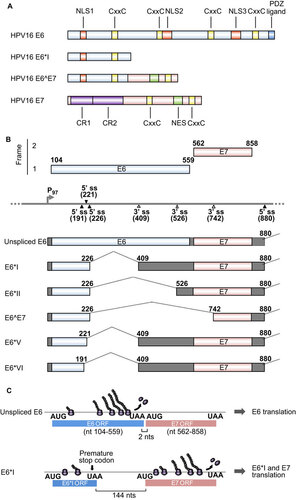
Figure 2 MCV T antigens and alternative RNA splicing. (A) Functional domains and motifs are indicated for the MCV large T antigen (LT), 57 kDa T antigen (57kT) and small T antigen (sT). (B) Scheme of alternative splicing of MCV T antigens. T antigen ORFs created by alternative RNA splicing (dashed lines) are in gray and 5′ and 3′ untranslated regions are in dark gray. Nucleotide positions are numbered according to the MCV genomic sequence (GenBank: NC_010277.1).
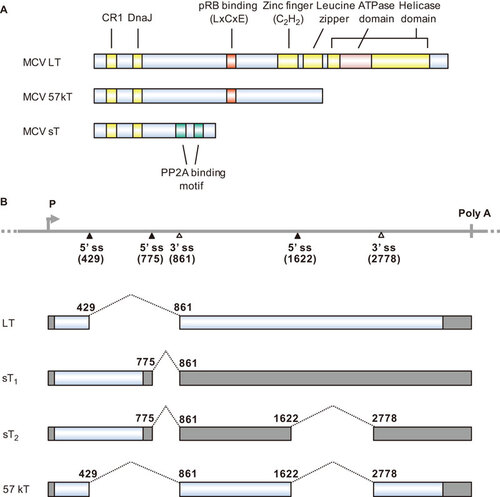
Figure 3 The EBV LMP1 oncoprotein and its lytic variant lyLMP1. (A) Functional domains and motifs are indicated for EBV LMP1 and lyLMP1 in the B95-8 EBV isolate. (B) EBV LMP1 ORF and lyLMP1 are produced by RNA splicing (dashed lines) from two separate transcripts derived either from the ED-L1 promoter or the ED-L1A promoter. Nucleotide positions are numbered according to the genomic DNA sequence of the B95-8 EBV isolate (GenBank: V01555.2). TRAF, TNF receptor-associated factor; TRADD, tumor necrosis factor receptor associated death domain protein.
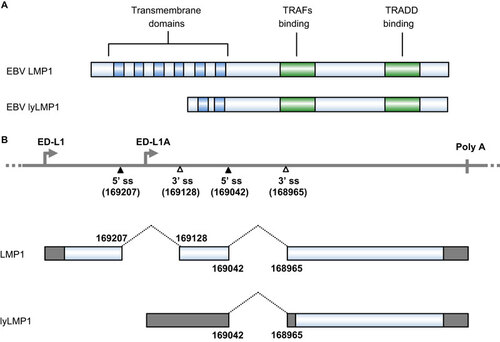
Figure 4 HTLV-1 viral RNA splicing and Tax oncogene expression. (A) Functional domains and motifs are indicated for the HTLV-1 Tax oncoprotein. (B) Alternative RNA splicing products of HTLV-1 viral genes. Seven positive-strand transcripts (1-E, 1-2-3, 1-2-A, 1-A, 1-2-B, 1-C and 1-3) and one negative strand transcript (HBZ) are produced by alternative RNA splicing of the transcripts derived from either a 5′ or 3′ long terminal repeat (LTR). The HTLV-1 Tax ORF in blue is present in the 1–2–3 mRNA, which is spliced from the nt 119 5′ ss to the nt 4641 3′ ss and then from the nt 4831 5′ ss to the nt 6950 3′ ss. Nucleotide positions are numbered according to the HTLV-1 genomic RNA sequence, starting from the first nucleotide in the pre-mRNA.Citation153
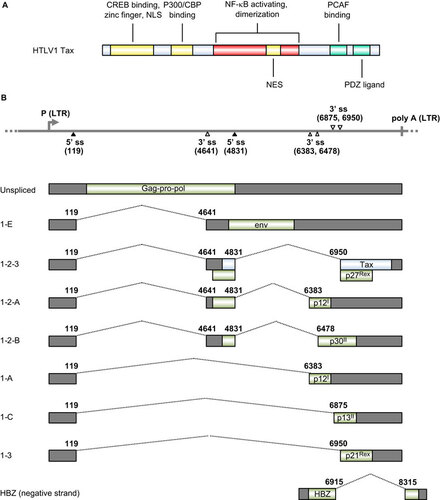
Figure 5 Transcription map of KSHV ORF73 (LANA), ORF72 (v-Cyclin), ORF71 (K13, v-FLIP) and K12 (kaposin). The diagram shows major alternative splicing products of KSHV ORF73, ORF72, ORF71 and K12. The heavy line indicates the corresponding latent locus region of KSHV genome. Three promoters (LTc, constitutive promoter; LTi, RTA-inducible promoter; LTd, downstream promoter), two polyadenylation signals (poly A at nt 122094 and nt 117432) and alternative 5′ ss and 3′ ss in this region are indicated. Twelve viral miRNAs (miR-K1-12) clustering downstream of the K13 ORF are shown as orange triangles. Boxes above the line represent ORFs for ORF73, ORF72, K13 and K12. Below the line are common RNA species (A–G) derived from alternative RNA splicing from this region. Nucleotide positions are indicated according to KSHV genome (GenBank: U75698.1).Citation181
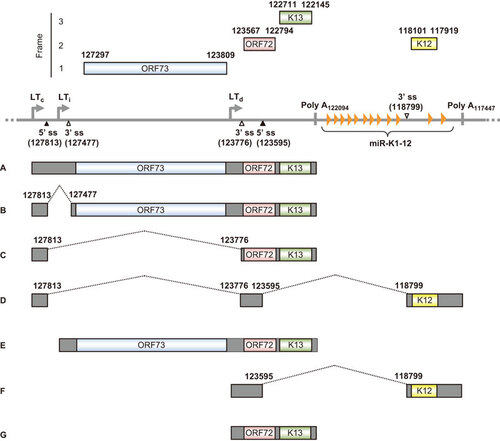
Figure 6 Genome structure and transcription map of HBV. The full-length, circular HBV genome (GenBank: X02496.1)Citation222 is illustrated in a linear form for better presentation of tail-to-head (3182/1) junction, four promoters (preC/pg, preS1, preS2 and X), a single poly A site at nt 1919, four alternative 5′ ss (filled triangles) and six alternative 3′ ss (open triangles). Above the linear HBV genome are viral ORFs, each with the numbered positions of the first nt of the start codon (including the in-frame start codon) and the last nt of the stop codon. Below the linear HBV genome are the RNA species (A–M) commonly derived from alternative RNA splicing, with the coding exons in colored boxes and the non-coding exons in grey boxes. The dotted lines indicate the introns and splicing directions for each RNA species, with the mapped splice site positions being numbered by nt positions in the HBV genome. HBSP, HBDSP and P-S FP are the ORFs created by alternative splicing of the preC/pg RNA. HBSP, hepatitis B splice-generated protein; HBDSP, hepatitis B doubly spliced protein; P-S FP, polymerase-surface fusion protein.