Figures & data
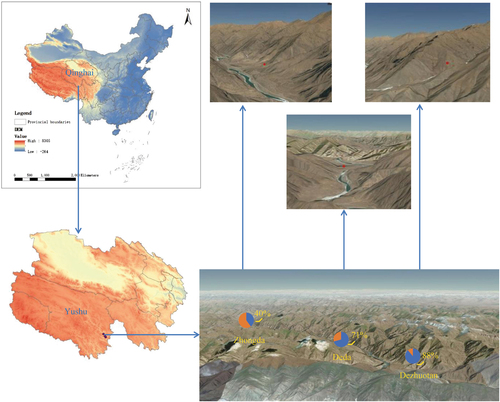
Red dots are the collection sites. Positive rates in three locations are indicated
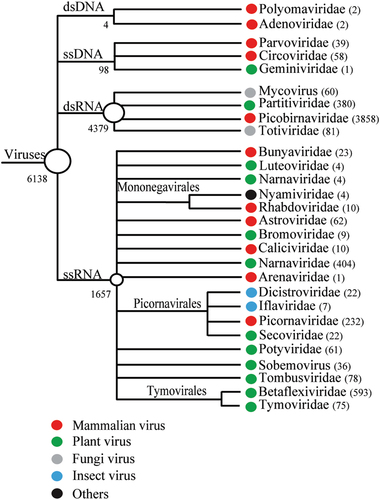
Numbers of virus contigs are shown in parentheses
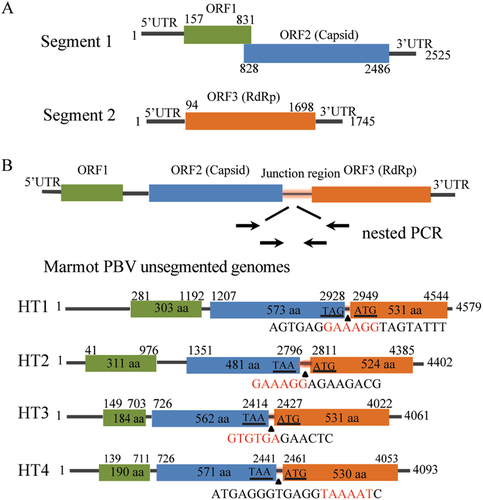
(a) Schematic representation of the two segments of human picobirnavirus strain Hy005102 (GenBank accession no. NC_007026 and NC_007027). (B) The unsegmented genomes of picobirnaviruses detected in marmot. Arrows indicate the position and direction of primers for nested PCR. Stop codon of the capsid gene and the initiation codon of the RdRp are underlined. Junction sequences between segments 1 and 2 of bi-segmented picobirnaviruses are shown below the triangles. Segmentation-associated motifs are shown in red
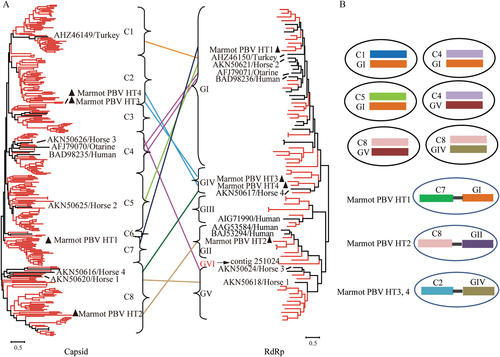
a The phylogenetic trees of the full-sequence capsid (left) and RdRp (right) proteins. Sequences of picobirnaviruses obtained from marmot are shown in red. Sequences of picobirnaviruses reported previously are shown in black. Unsegmented picobirnaviruses from marmot are indicated by black triangles. Capsid and RdRp sequences of picobirnaviruses that underwent assortment are linked by lines. b Proposed assortment types of bi-segmented and unsegmented picobirnaviruses
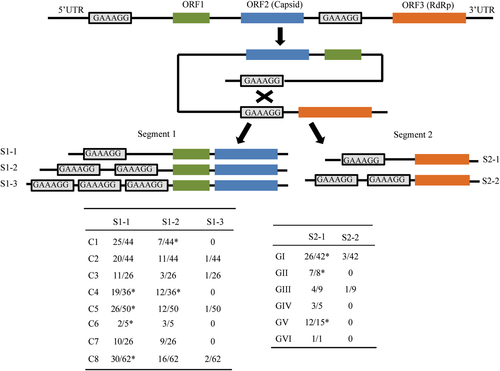
Relative positions of the 6-bp direct repeat sequence GAAAGG in the 5′ UTR and junction between the capsid and RdRp genes of unsegmented Marmot picobirnaviruses are indicated by rectangles. A schematic of the three types of segment 1 harboring GAAAGG (S1-1-3) and two types of segment 2 harboring GAAAGG (S2-1-2) from bi-segmented picobirnaviruses. The proportions of S1-1-3 and S2-1-2 among the total sequences analyzed, together with their genotypes, are shown in the following tables. *The published segment 1 sequences (GenBank accession no. KJ495689; KF861768; JQ776551; AB186897; KR902506; KC692367; KR902502; KR902504; KF861770; KF861771; KF861772). The published segment 2 sequences (GenBank accession no. JQ776552; AAG53583; AHX00960; ACT64131; AFK81927; BAJ53294; AHZ46150; KR902507; KR902503)