Figures & data
Minimum inhibitory concentrations of tested antimicrobial agents for the studied bacterial isolates
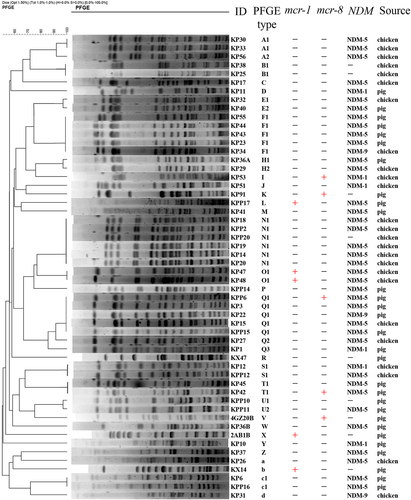
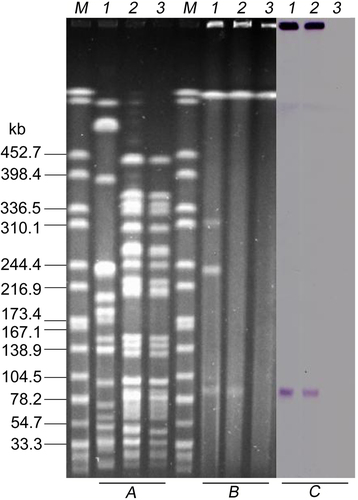
aXbaI-digested PFGE of K. pneumoniae KP91, transconjugants, and the recipient E. coli strain J53. b S1-PFGE and c the corresponding Southern hybridization using the mcr-8-specific probe. Lane M, marker H9812; lane 1, K. pneumoniae KP91; lane 2, transconjugant E. coli J53-pKP91; lane 3, recipient E. coli J53
Circles display (outside to inside) (i) size in bp and (ii) the positions of predicted coding sequences transcribed in a clockwise orientation
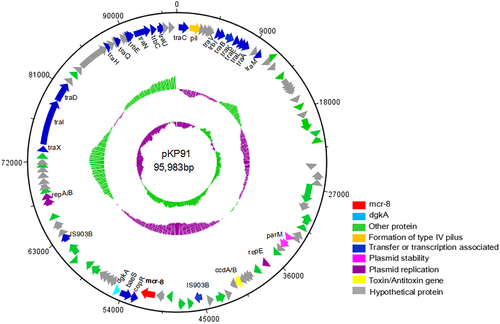
A logo represents each column of the alignment in a stack of letters, with the height of each letter proportional to the observed frequency of the corresponding amino acid, and the overall height of each stack proportional to the sequence conservation, measured in bits, at that position. Red asterisks indicate the six conserved cysteine residues that form three disulfide bonds. Black asterisks indicate the conserved active sites among MCR-1–8. The logo was built using WebLogo software (http://weblogo.berkeley.edu/logo.cgi)
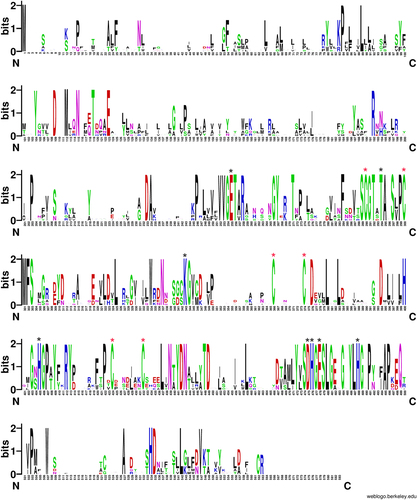
a Structure prediction for MCR-8 and reference protein MCR-3. Domain 1 was predicted to be a transmembrane domain, while domain 2 was predicted to be a phosphoethanolamine transferase. b The five transmembrane α-helices predicted by the Philius transmembrane prediction server (type confidence, 0.99; topology confidence, 0.88)
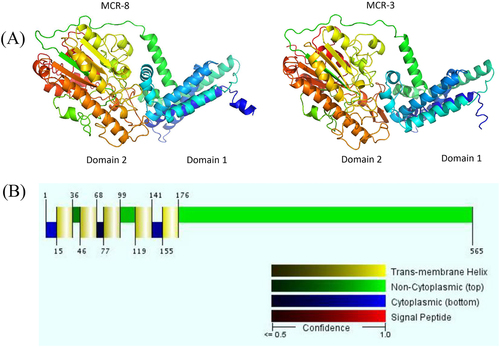
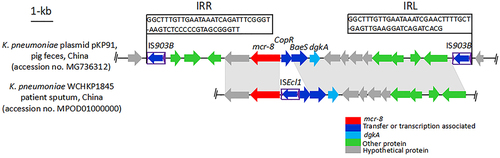
K. pneumoniae WCHKP1845 was isolated from the sputum of patients in China (accession no. MPOD01000000). The positions and orientations of the genes are indicated by arrows, with the direction of transcription shown by the arrowhead. Gray shading indicates > 90% nucleotide sequence identity