Figures & data
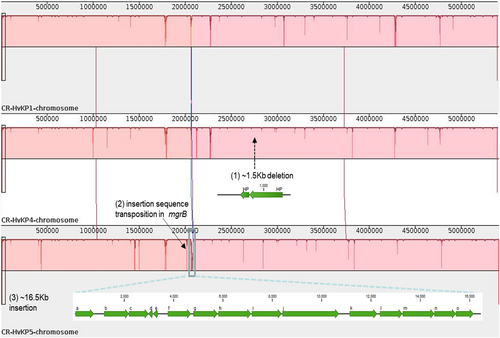
List of chromosomal SNP differences among the clonal CR-hvKP outbreak strains tested
Major regions of divergence are labeled, including: (1) An 1.5-Kb deletion in the K. pneumoniae 4 genome with two genes encoding hypothetical proteins (indicated with a black arrow); (2) disruption of the mgrB gene by insertion sequence (ISKpn18) in the K. pneumoniae 5 genome (indicated with a black arrow); (3) A 16.5-Kb region of the K. pneumoniae 5 genome (indicated by a blue rectangle) is absent in the other two ST11 K. pneumoniae genomes. Predicted genes within this region are indicated: (a) peptidase; (b) fatty acid desaturase; (c) phosphate ABC transporter substrate-binding protein; (d) nitrilotriacetate monooxygenase; (e) hypothetical protein; (f) transcriptional regulator TdcA; (g) bifunctional threonine ammonia-lyase/L-serine ammonia-lyase TdcB; (h) threonine/serine transporter TdcC; (i) propionate kinase; (j) formate C-acetyltransferase; (k) branched-chain amino acid ABC transporter substrate-binding protein; (l) branched-chain amino acid ABC transporter permease, LivH; (m) branched-chain amino acid ABC transporter permease, LivM; (n) ABC transporter ATP-binding protein, LivG; (o) ABC transporter ATP-binding protein, LivF
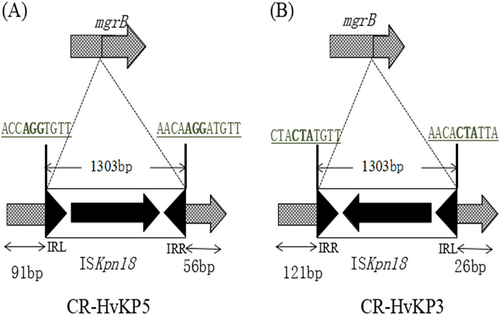
The mgrB gene was truncated by ISKpn18 in both K. pneumoniae 5 (a) and K. pneumoniae 3 (b) strains with opposite directions at different positions. Target site duplications are underlined with bold letters. The left and right inverted repeats (IRL and IRR) of ISKpn18 are represented as black triangles
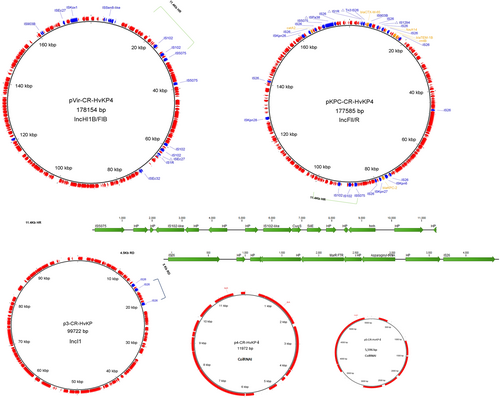
Each of the three ST11 isolates carried five plasmids, with sizes of 178, 177.5, 99.7, 11.9, and 5.6 Kb. Insertion sequences and antimicrobial resistance genes are annotated with blue and yellow fonts, respectively. Plasmid name, size, and replicon types are depicted in the middle of each circle. The 11.4-Kb homologous region (HR) in the virulence plasmid and MDR plasmid, and 4.5-Kb region of divergence (RD) in the 99.7-Kb plasmid are highlighted